Abstract
Background
The respiratory tract harbors a variety of microbiota, whose composition and abundance depend on specific site factors, interaction with external factors, and disease. The aim of this study was to investigate the relationship between COVID-19 severity and the nasopharyngeal microbiome.
Methods
We conducted a prospective cohort study in Mexico City, collecting nasopharyngeal swabs from 30 COVID-19 patients and 14 healthy volunteers. Microbiome profiling was performed using 16S rRNA gene analysis. Taxonomic assignment, classification, diversity analysis, core microbiome analysis, and statistical analysis were conducted using R packages.
Results
The microbiome data analysis revealed taxonomic shifts within the nasopharyngeal microbiome in severe COVID-19. Particularly, we observed a significant reduction in the relative abundance of Lawsonella and Cutibacterium genera in critically ill COVID-19 patients (p < 0.001). In contrast, these patients exhibited a marked enrichment of Streptococcus, Actinomyces, Peptostreptococcus, Atopobium, Granulicatella, Mogibacterium, Veillonella, Prevotella_7, Rothia, Gemella, Alloprevotella, and Solobacterium genera (p < 0.01). Analysis of the core microbiome across all samples consistently identified the presence of Staphylococcus, Corynebacterium, and Streptococcus.
Conclusions
Our study suggests that the disruption of physicochemical conditions and barriers resulting from inflammatory processes and the intubation procedure in critically ill COVID-19 patients may facilitate the colonization and invasion of the nasopharynx by oral microorganisms.
Keywords: Nasopharyngeal microbiome, Nasopharynx, COVID-19, Staphylococcus, Corynebacterium, Streptococcus, Lawsonella, Cutibacterium Core microbiome, Intubation
Highlights
-
•
Invasion of the oral microbiota to the nasopharynx in Critical COVID-19.
-
•
Intubation can have a discernible impact on the nasopharyngeal microbiome.
-
•
Core Nasopharyngeal Microbiome: Corynebacterium, Staphylococcus, and Streptococcus.
1. Introduction
Respiratory tract is characterized by unique anatomical structures, performing various functions and harboring distinct microenvironments. The nasal cavity, nasopharynx, and oropharynx are part of the upper respiratory tract (URT). Despite their anatomical proximity, these structures exhibit differences in their microbiota. Genera such as Staphylococcus, Propionibacterium, Cutibacterium, Corynebacterium, Moraxella, and Streptococcus have been identified as predominant in the nasal cavity [1,2]. Nasopharynx, with its unique microenvironment characterized by specific pH, temperature, relative humidity, and the presence of pharyngeal tonsils, is permissive to the colonization by different microorganisms, including Dolosigranulum, Neisseria, and Haemophilus [3]. Due to the proximity of its connection with the mouth, the oropharynx harbors a distinct microbial community that includes Rothia, Veillonella, Prevotella, Atopobium, Gemella, and Streptococcus, amongst others [4]. This structure has the highest bacterial load in the respiratory tract, with many microorganisms originating from oral microbiota [5].
It is well known that respiratory microbiome is a complex ecosystem, comprising diverse microorganisms that participate in cooperative and competitive interactions among themselves, and with the host, thereby impacting respiratory health and disease. The coexistence of two microbial species in the same niche from airways ecosystems, with identical needs usually is not feasible [6]. Each anatomical site within the respiratory tract signifies a distinct niche influenced by interactions with the host immune system and diverse external factors. Respiratory viral infections, provoke changes in the physicochemical and immunological conditions of the epithelial barrier that impact in microbial competition for specific ecological niches [7], leading to modifications in microbial colonization. In this context, COVID-19, a highly infectious respiratory disease caused by the SARS-CoV-2 virus, causes inflammation in the respiratory tract, and in severe cases, respiratory tract damage may progress to respiratory failure and acute respiratory distress syndrome (ARDS) [[8], [9], [10]] triggering pathogenic immune response associated to epithelial respiratory tract and lung tissue damage [11,12]. These inflammatory and physicochemical changes in the mucosal membranes may provoke the translocation of microorganisms from different compartments, such as the gut to the blood [13] or the mouth to the respiratory tract [14]. This phenomenon could be a cause of secondary infections or co-infections by microorganisms such as Staphylococcus, Streptococcus, Pseudomonas, Klebsiella, and Acinetobacter, thereby contributing to the severity of COVID-19 [15,16]. These complications often result in extended hospital stays and can even lead to fatal outcomes [17]. Previous studies have focused on comparing the diversity of the nasopharyngeal microbiome in COVID-19 [18,19]. Prasad et al. [20] and Mostafa et al. [21] reported a decrease in alpha diversity, while Braun et al. [18] and Feehan et al. [22] did not find significant differences in alpha diversity indices. Taxonomic changes have been reported in the relative abundance of Corynebacterium, Dolosigranulum, Staphylococcus, Fusobacterium, Anaerococcus, Enterobacter, Peptostreptococcus, Prevotella, Pseudomonas, Mycoplasma, Alloprevotella, Solobacterium, Veillonella, Streptococcus, and Actinomyces genera in COVID-19 patients compared to healthy individuals [15,[22], [23], [24], [25]]. In general, the results of these studies are still non consistent.
In this study, the relationship between the severity of COVID-19 and the nasopharyngeal microbiome was evaluated. Thus, we analyzed the bacterial microbiome of patients with different degree of COVID-19 disease severity and compared them to healthy volunteers. Our analysis, along with the identification of the core microbiome and the changes in nasopharyngeal genera, might contribute to our understanding of microbial interactions in patients with different clinical presentations of COVID-19.
2. Materials and methods
2.1. Study population
We conducted a prospective cohort study in 44 adult subjects, including 30 SARS-CoV-2 positive individuals by real-time PCR and 14 healthy volunteers negative for SARS-CoV-2 infection. Patients were recruited from the emergency room unit at the Instituto Nacional de Enfermedades Respiratorias “Ismael Cosío Villegas” (INER) in Mexico City, which is a referral center for respiratory diseases in Mexico. Nasopharyngeal swabs were collected from fall 2020 to fall 2021 and only patients with positive real-time PCR analysis for SARS-CoV-2 were recruited. Samples from healthy donors were collected in parallel. Healthy volunteers had no respiratory symptoms and were negative for the RT-PCR test, at the time of sample collection and for 4 weeks thereafter. COVID-19 severity was categorized according to WHO's Living Guideline Clinical Management of COVID-19 (January 13th, 2023), 9 patients had severe COVID-19 disease and 21 were classified as critically ill requiring mechanical ventilation. The list of the samples and clinical and demographic information of patients are summarized in Supplementary Table 1.
2.2. Sample processing
Nasopharyngeal swab samples were taken according to the standard guidelines. The sample was placed in UTM®: Universal Transport Medium™ (Copan Diagnostics, United States) and stored at −70°C until its DNA extraction was performed. Samples were centrifuged for 20 min at 14,000 RPM, the supernatant was removed and discarded. The pellet was resuspended in 500 μl of Sputolysin® (Calbiochem, Germany) and was mixed on a vortex mixer for 30 s, the suspended mixture stood at room temperature for 15 min. Subsequently, it was centrifuged for 5 min at 1500 rpm, discarding the supernatant. DNA was extracted with the QIAmp UCP pathogen miniKit (Qiagen), using the mechanical pre-lysis with spin protocol for swabs described in the QIAmp UCP Pathogen Mini Handbook. The incubation time with Proteinase K was performed during 20 min. Subsequently, the DNA V4 region of 16S rRNA was amplified by PCR using the primers 515f and 806r [26]. Samples were pooled and barcoded following Illumina 16S metagenomics protocol (Illumina, USA). The barcodes were pooled in equimolar concentration and sequenced on the Illumina Miseq platform using the 2 × 250 pair-end method.
2.3. Bioinformatics analysis and statistics
Sequencing data was processed with R version 4.2.1. Quality trimming, sample inference, paired reads merging, chimera removal, and taxonomy assignment were performed using DADA2 R package version 3.17 [27]. Silva v138 database was utilized for taxonomic assignment. ASV abundances were normalized using the Wrench method [28]. The analysis of alpha diversity was performed using the Microbiome R package version 1.18 [29]. For beta diversity analysis Non-metric multidimensional scaling (NMDS) was used, using the Bray-Curtis dissimilarity index, and permanova analysis ADONIS2 was used to determine significance. Statistical tests and charts were performed using Ggstatsplot R package version 0.9.4 [30] and Gtsummary R package version 1.6.2 [31] to evaluate the differences between groups and demographic characteristics, comorbidities, clinical data, laboratory tests, severity of illness, and clinical outcome. The core microbiome was defined as taxonomic groups that were present in at least 60 % of the samples with 0.001 % relative abundance, using the Eulerr, Microbiome, and Microbiomeutilities R packages. Correlation analysis was performed to explore the relationships between variables of interest. Spearman's rank correlation coefficient was utilized to assess the strength and direction of associations between the relative abundances of taxonomic groups and severity of COVID-19. The severity of COVID-19 was categorized into three ordinal levels: controls (assigned a value of 0), moderate COVID-19 cases (assigned a value of 1), and critical COVID-19 cases (assigned a value of 2). Correlation analysis was conducted using statistical software R with significance set at p < 0.05.
Additionally, we conducted exploratory data analysis to assess the impact of covariates, including comorbidities, antibiotic usage, and steroid administration prior to sampling, further the effect of orotracheal intubation for mechanical ventilation. Our analysis involved a comparative examination of each covariate using the relative abundances of taxonomic groups. To conduct this analysis, we employed Mann-Whitney statistical tests, utilizing the Ggstatsplot R package (version 0.9.4) and the Gtsummary R package (version 1.6.2).
2.4. Bioinformatic analyses based in databases reported in other studies
Furthermore, to strengthen our analysis, a systematic search was conducted in PubMed of the National Center for Biotechnology Information (NCBI) to identify studies focusing on the nasopharyngeal microbiota and COVID-19 severity. A total of six relevant studies were identified, all of which included healthy volunteers and employed high-throughput amplicon sequencing based on the 16S rRNA gene to analyze the microbiome. After reviewing these six studies, we found that only two of them had accessible data on samples from the nasopharynx in both COVID-19 patients and healthy volunteers. Specifically, Smith et al. [25] conducted their study in France, while Hurst et al. [32] conducted an interesting study on patients from the United States. Both studies provided complete metadata and available sequences, enabling us to examine nasopharyngeal samples across varying severities of COVID-19 and compare them to those from healthy volunteers. Thus, in order to determine differences in microbial abundance in patients and controls and compare their reports with our data, we processed the data obtained from these studies. The sequencing data and meta data were downloaded from the SRA database for projects PRJNA714242 and PRJNA703574. Subsequent preprocessing steps involved quality control measures, including the removal of adapter sequences, elimination of low-quality reads, and filtering out sequences containing ambiguous bases using the DADA2 package in R. For Smith et al. [25], sequences, we used the filterAndTrim parameters truncLen = c(262,230), maxN = 0, maxEE = c(2,2), truncQ = 2, and rm. phix = TRUE. For Hurst et al. [32], we used the filter parameters truncLen = c(150,160), maxN = 0, maxEE = c(2,2), truncQ = 2, and rm. phix = TRUE. Following preprocessing, the sequences underwent error correction, dereplication, merging of paired reads, identification of unique sequence variants, and taxonomy annotation using our bioinformatic pipeline.
For more specific details see Supplementary Materials and Methods.
3. Results
A total of 44 subjects were included in the study, all of them were adults residing in Mexico, with a median age of 49 (42–61) years, and 16 (36 %) were female. Body Mass Index (BMI) categories were as follows: 5 (11 %) had normal weight, 19 (43 %) were overweight, 16 (36 %) presented obesity, and 4 (9.1 %) BMI was not reported. The studied groups were classified according to COVID-19 severity. Severe and critical COVID-19 patients were more frequently obese (p = 0.002). We found a statistically significant difference between the groups in terms of medical history related to tobacco use (p = 0.03) (Table 1). Fever, cough, dyspnea, arthralgias, myalgias, and decreased SpO2 were the most prevalent symptoms in COVID-19 patients. We found decreased levels of lymphocytes (p = 0.012) and high levels of neutrophils (p = 0.010) in COVID-19 patients when compared to controls. The neutrophil-lymphocyte ratio (NLR) was statistically significantly different between groups (p = 0.001).
Table 1.
Demographic characteristics and comorbidities.
| Control, N = 14 | Severe, N = 9 | Critical, N = 21 | P-value* | |
|---|---|---|---|---|
| Age | 52 (40, 60) | 47 (47, 55) | 48 (42, 65) | 0.8 |
| Gender | >0.9 | |||
| female | 6 (43 %) | 3 (33 %) | 7 (33 %) | |
| male | 8 (57 %) | 6 (67 %) | 14 (67 %) | |
| Height | 1.68 (1.54, 1.75) | 1.67 (1.64, 1.68) | 1.70 (1.60, 1.73) | 0.9 |
| Unknown | 4 | 0 | 0 | |
| Weight | 72 (62, 81) | 82 (74, 85) | 80 (75, 91) | 0.14 |
| Unknown | 4 | 0 | 0 | |
| BMI categories | 0.002 | |||
| Normal weight | 2 (14 %) | 1 (11 %) | 2 (9.5 %) | |
| Obesity | 0 (0 %) | 5 (56 %) | 11 (52 %) | |
| Overweight | 8 (57 %) | 3 (33 %) | 8 (38 %) | |
| No data | 4 (29 %) | 0 (0 %) | 0 (0 %) | |
| Comorbidities | ||||
| Obesity | 1 (7.1 %) | 0 (0 %) | 8 (38 %) | 0.031 |
| DM2 | 1 (7.1 %) | 1 (11 %) | 6 (29 %) | 0.3 |
| Hypertension | 1 (7.1 %) | 3 (33 %) | 9 (43 %) | 0.066 |
| Heart disease | 0 (0 %) | 0 (0 %) | 1 (4.8 %) | >0.9 |
| Renal insufficiency | 0 (0 %) | 0 (0 %) | 1 (4.8 %) | >0.9 |
| Immunosuppression | 0 (0 %) | 0 (0 %) | 0 (0 %) | |
| Asthma | 1 (7.1 %) | 1 (11 %) | 0 (0 %) | 0.3 |
| COPD | 0 (0 %) | 0 (0 %) | 0 (0 %) | |
| HIV | 0 (0 %) | 0 (0 %) | 0 (0 %) | |
| GERD | 2 (14 %) | 1 (11 %) | 0 (0 %) | 0.2 |
| Allergic rhinitis | 0 (0 %) | 2 (22 %) | 0 (0 %) | 0.038 |
| Alcoholism | 0 (0 %) | 2 (22 %) | 5 (24 %) | 0.12 |
| Smoking | 0 (0 %) | 1 (11 %) | 7 (33 %) | 0.031 |
| Initial Antibiotic | 0 | 8 (89 %) | 15 (71 %) | <0.001 |
| Days hospitalized | 0 (0, 0) | 11 (0, 16) | 29 (13, 37) | |
| Days onset sampling | 0 (0, 0) | 9 (7, 12) | 10 (8, 14) | |
Median (IQR); n (%); *Kruskal-Wallis rank sum test; Fisher's exact test.
BMI Body Mass Index, DM2 Diabetes Mellitus Type 2, GERD Gastroesophageal reflux disease.
3.1. Respiratory microbiome composition in COVID-19 patients with different clinical presentations
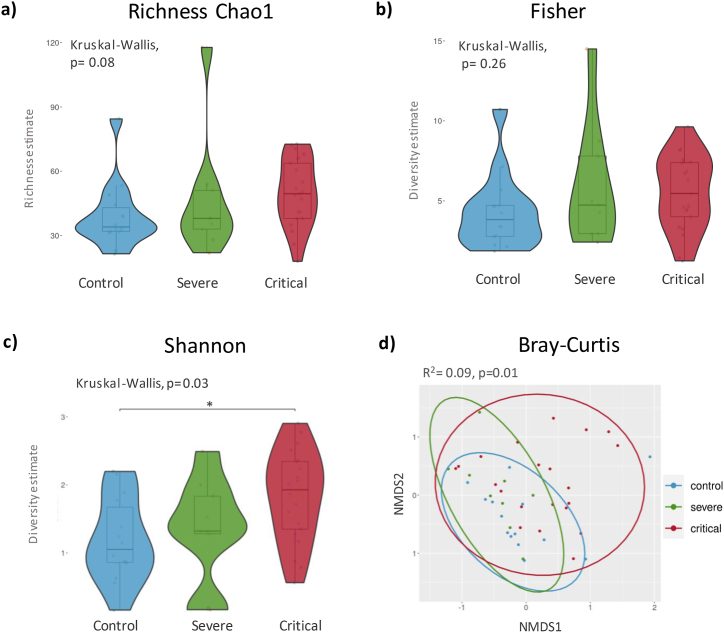
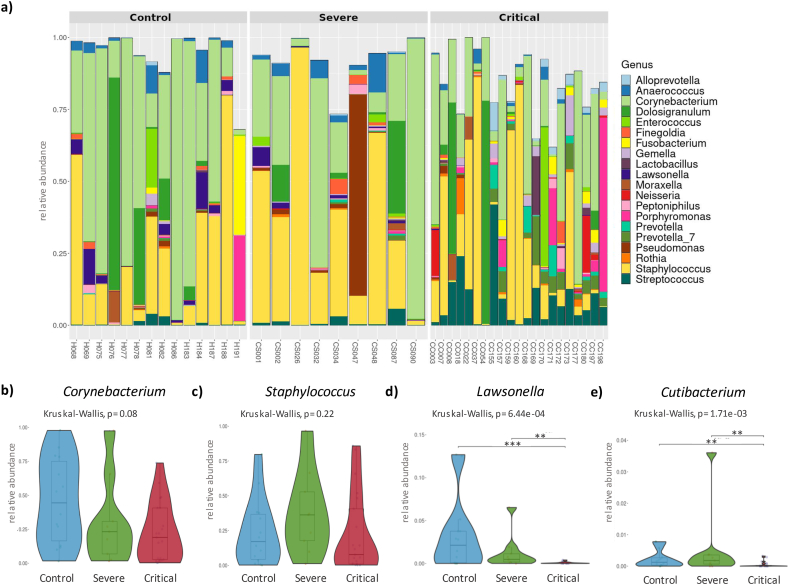
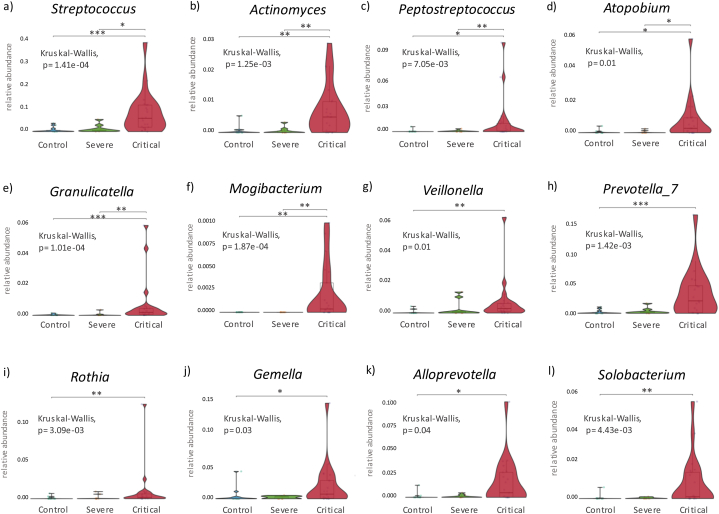
The abundance of amplicon sequence variants (ASV) at the phylum level, revealed an increase in Bacteroidota (p = 0.010) and Fusobacteria (p = 0.026) in samples from COVID-19 patients. Actinobacteria and Firmicutes phyla were the most abundant in COVID-19 and healthy controls but showed no significant differences amongst these groups. Shannon index showed an increase in the bacterial diversity in critically ill COVID-19 patients (p = 0.03) (Fig. 1c) however the remaining alpha indexes did not show significant differences between critically ill and severe COVID-19 patients and controls (Fig. 1a and b). Non-metric dimensional scaling (NMDS) was used to compare beta diversity and revealed significant differences between critically ill COVID-19 patients and controls (R2 = 0.09 p = 0.01) (Fig. 1d). At genus level, Corynebacterium (0.24 [0.06, 0.50]) and Staphylococcus (0.14 [0.03, 0.39]) were the most abundant (Fig. 2a), with no significant differences between groups (Fig. 2b and c). Importantly, the presence of the genera Lawsonella and Cutibacterium was low in severe and critically ill COVID-19 patients (p < 0.001) (Fig. 2d and e). Contrariwise, Streptococcus, Actinomyces, Peptostreptococcus, Atopobium, Granulicatella, and Mogibacterium (Fig. 3 a-f) showed an increase in severe and critical COVID-19 patients (p < 0.01). Other genera that exhibited increased abundance only in critical COVID-19 compared to controls were Veillonella, Prevotella_7, Rothia, Gemella, Alloprevotella, and Solobacterium (p < 0.01) (Fig. 3 g-l).
Fig. 1.
Richness and alpha diversity indices. a) Richness Chao1 index, b) Fisher index showing no significant differences. c) Shannon index revealing a significant difference between the control and critical COVID-19 (p = 0.03). d) Beta diversity of the nasopharyngeal microbiome among different severity groups of COVID-19. Significance was determined using Kruskal-Wallis test with Dunn's multiple comparison with a 95 % confidence interval. The NMDS plot is based on Bray-Curtis dissimilarity, and PERMANOVA statistics indicate a significant difference among the three groups. Each color represents a specific analyzed group. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Fig. 2.
a) Taxonomy bar plot showing the top 20 microbiome profiles at the genus level, ranked by relative abundance, in control, severe, and critical patients. b) and c) Violin plot displaying the relative abundance of Staphylococcus and Corynebacterium genera. No significant differences were observed between severity groups. c) and d) Decrease in the relative abundance of Lawsonella and Cutibacterium genera in critical COVID-19 patients. Significance was determined using the Kruskal-Wallis test with Dunn's multiple comparison, 95 % confidence interval, where ∗ p ≤ 0.05, ∗∗ p ≤ 0.01, and ∗∗∗ p ≤ 0.001.
Fig. 3.
Genera with increased relative abundance in critical COVID-19 patients. Significance was determined using the Kruskal-Wallis test with Dunn's multiple comparison 95 % confidence interval, where ∗ p ≤ 0.05, ∗∗ p ≤ 0.01, and ∗∗∗ p ≤ 0.001.
To assess the strength and direction of the association between different genera and the severity of COVID-19 illness, a Spearman rank correlation analysis was done. We observed moderate to strong negative correlations of Corynebacterium (rho = −0.334, p-value = 0.026), Cutibacterium (rho = −0.509, p-value = 0.0004), and Lawsonella (rho = −0.562, p-value = 7.192e-05) with COVID-19 severity. In Contrast, Streptococcus exhibited a strong positive correlation with COVID severity (rho = 0.637, p-value = 3.315e-06), while several other genera including Prevotella, Actinomyces, Solobacterium, Atopobium, Mogibacterium, Alloprevotella, Veillonella, Rothia, Granulicatella, and Peptostreptococcus demonstrated moderate positive correlations with COVID-19 severity (Supplementary Fig. 1).
3.2. Analysis of confounding covariates
The analysis of antibiotic use before sampling revealed a statistically significant effect (p < 0.05) among the genera Prevotella 7, Alloprevotella, Rothia, and Granulicatella between the control group and COVID-19 patients. Regarding comorbidities, only Solobacterium showed statistical significance (p = 0.03). However, the previous administration of steroids demonstrated a significant effect across almost all genera of interest in this study (p < 0.05) (Supplementary Table 2). Notably, Staphylococcus and Cutibacterium did not exhibit significant differences based on steroid administration.
3.3. Microbiome composition after intubation
Since previous studies have indicated that the use of invasive respiratory devices can disrupt the microenvironment and alter microbial populations [33], we decided to investigate whether such changes occurred in our studied groups of patients. For this analysis, we divided the group of critical patients into two subgroups: Pre-intubation and Post-Intubation, based on the timing of sample collection about the time of intubation for mechanical ventilatory support. The first subgroup comprised patients sampled before intubation [n = 7], with an average time of 1.85 days (range 0–5 days) between sample collection and orotracheal intubation. The second subgroup consisted of patients sampled after intubation (n = 14), with an average time of 2 days (range 0–7 days) between orotracheal intubation and sample collection. It is crucial to note that each subgroup consists of different patients. For additional details about the critical patient's intubation time line refer to Supplementary Fig. 2. Based on this analysis we found a statistically significant increase in the alpha diversity indices: Chao1 (p = 0.001), Fisher (p = 0.03), and Shannon (p = 0.05) (Supplementary Figs. 3a, 3b, and 3c) among critically ill COVID-19 patients sampled before and after receiving mechanical ventilation support. Furthermore, beta diversity analysis using the Bray-Curtis index demonstrated significant differences between the Pre-intubation and Post-intubation samples (R2 = 0.08, p = 0.03) (Supplementary Fig. 3d). Analysis of the genus composition after intubation revealed that the distribution of Corynebacterium, Staphylococcus, Lawsonella, Cutibacterium, Streptococcus, Veillonella, Rothia, and Granulicatella was not different between the two groups. Interestingly, Prevotella_7, Atopobium (p < 0.001), Solobacterium (p = 0.003), and Actinomyces (p = 0.008), exhibited an increased abundance after placement of the orotracheal tube (Supplementary Fig. 4).
3.4. Nasopharyngeal core microbiome
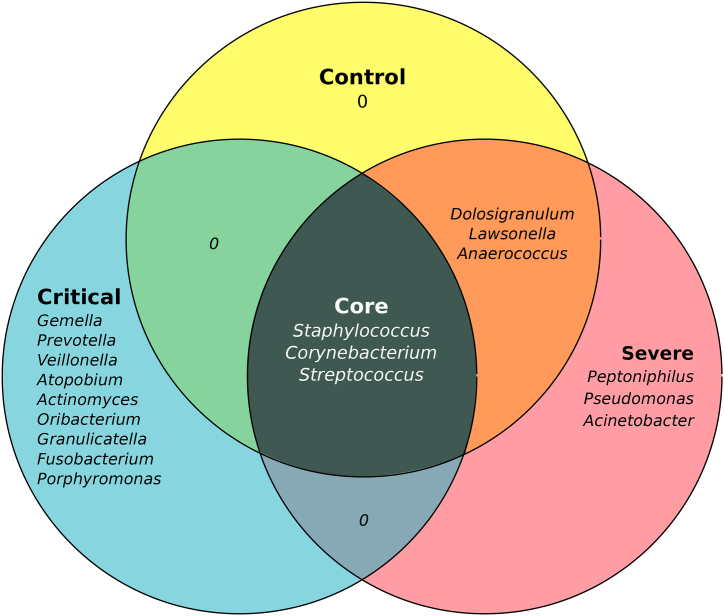
One of the challenges in microbiome research is to identify microorganisms that are consistently and stably present in most samples, within a specific niche. In our analysis, the common core of the microbes presents in the analyzed samples, consisted of the genera Staphylococcus, Corynebacterium, and Streptococcus. Healthy controls and COVID-19 severe cases shared the genera Dolosigranulum, Lawsonella, and Anaerococcus. Regardless of this, additional bacterial genera Peptoniphilus, Pseudomonas, and Acinetobacter were identified in COVID-19 severe cases. While critically ill patients showed an enrichment of the genera Gemella, Prevotella, Veillonella, Atopobium, Granulicatella, Actinomyces, Oribacterium, Fusobacterium, and Porphyromonas (Fig. 4).
Fig. 4.
Nasopharyngeal Core Microbiome. Microorganisms shared across communities, are present in at least 60 % of the samples with 0.001 % of relative abundance.
3.5. Insights from other studies of nasopharyngeal microbiome in COVID-19 patients
After the COVID-19 epidemic, people started looking into how the body's microbiome affects the disease. As a result, we identified different studies with accessible raw data, obtained from nasopharynx samples from COVID-19 patients and healthy volunteers [25,32]. From these studies, we selected and downloaded raw data to perform comparative bioinformatic analyses with our data. In the analysis that we performed from the data published by Smith et al. [25], we observed an increase in the abundance of the genera Staphylococcus and Veillonella, accompanied by a decrease in Corynebacterium, Dolosigranulum, and Lawsonella among critically ill COVID-19 patients. In contrast, the study conducted by Hurst et al. [32], did not reveal statistically significant decreases in any genera between severity groups and healthy volunteers; only an increase in Fusobacterium was noted in moderate COVID-19 patients. Importantly, Corynebacterium, Staphylococcus, and Streptococcus were consistently present in all samples from healthy volunteers across both studies (Table 2).
Table 2.
| Comparison with other previously published studies.
| Data Set | Smith et al. [25] | Hurst et al. [32] | This study |
|---|---|---|---|
| COVID-19 asymptomatic | 0 | 5 | 0 |
| COVID-19 moderate patients | 15 | 22 | 0 |
| COVID-19 severe patients | 11 | 0 | 9 |
| COVID-19 critical patients | 23 | 0 | 21 |
| Healthy volunteers | 12 | 4 | 14 |
| Total Subjects | 61 | 31 | 44 |
| Source | Nasopharyngeal | Nasopharyngeal | Nasopharyngeal |
| Geography | France | USA | Mexico |
| Year | 2021 | 2022 | 2023 |
| Analysis | 16S rRNA gene, V3–V4 | 16S rRNA gene, V4 | 16S rRNA gene, V4 |
| Genus with Increased Relative Abundance by COVID Severity | Staphylococcus, Veillonella | no significant difference | Streptococcus, Actinomyces, Prevotella_ 7, Peptostreptococcus, Veillonella, Rothia, Solobacterium, Atopobium, Granulicatella, Gemella, Mogibacterium, Alloprevotella |
| Genus with decreased relative abundance by COVID Severity | Corynebacterium, Dolosigranulum, Lawsonella | Fusobacterium | Lawsonella, Cutibacterium |
| Nasopharynx Core Microbiome in Healthy Individuals | Corynebacterium, Staphylococcus, Escherichia-Shigella, Methylobacterium- Methylorubrum, Acinetobacter, Streptococcus, Pseudomonas, Lawsonella | Corynebacterium, Staphylococcus, Dolosigranulum, Lawsonella, Anaerococcus, Streptococcus | Staphylococcus, Corynebacterium, Dolosigranulum, Streptococcus, Lawsonella, Anaerococcus, |
| Accession number | PRJNA714242 | PRJNA703574 | PRJNA981220 |
4. Discussion
Here, we conducted a comparative analysis of the diversity and composition of the nasopharyngeal microbiome to explore its potential relationship with COVID-19 severity. Also, using a taxonomic profiling approach, we identified core microbiome genera associated with the nasopharynx in both healthy volunteers and COVID-19 patients.
Our main findings were 1) alpha diversity indexes were not different between severe and critically ill COVID-19 patients and controls and 2) COVID-19 patients present significant changes in the relative abundance of genera Lawsonella and Cutibacterium that were diminished, whereas genera Streptococcus and Actinomyces amongst other genera from oral microbiota were increased in severe but particularly in critically ill patients.
In line with our results, previous studies performed in other populations also described homogeneous alpha indexes of diversity in COVID-19 severe patients [18,22,34]. In this regard, Feehan et al. [22], described that alpha diversity did not differ in COVID-19 independently of the clinical status, but interestingly the alpha index was modified by other factors such as age and tobacco use. Contrasting those findings Shilts et al. and Ventero et al. [35,36], reported a decrease in alpha diversity in critically ill COVID-19 patients or those with fatal outcomes.
Another relevant finding of our study was that genera Staphylococcus, Streptococcus, and Corynebacterium were present as a core in all individuals including controls and COVID-19 patients. Furthermore, we identified in COVID-19 severe cases genera Peptoniphilus, Pseudomonas, and Acinetobacter and critically ill patients an enrichment of the genera Gemella, Prevotella, Veillonella, Atopobium, Granulicatella, Actinomyces, Oribacterium, Fusobacterium, and Porphyromonas. The meta-analysis of Reubens et al. [37] regarding studies on the diversity of the upper respiratory tract (URT) microbiome is consistent with our results, in that they find a trend in COVID-19 infections showing a reduction in some bacterial genera and an increase in others. However, the inclusion of both nasopharyngeal and oropharyngeal samples in their meta-analysis is a limitation that could explain discrepancies between their study and ours such as the observed higher abundance of Cutibacterium in their population.
We meticulously considered the potential influence of covariates beyond COVID-19 severity. Despite the presence of these covariates, we were able to identify discernible alterations in taxonomic composition among critically ill COVID-19 patients, suggesting that Lawsonella and Cutibacterium decrease additionally to Streptococcus increase was independent of confusing factors such as comorbidities, intubation, and prior antibiotic intake. We also observed statistically significant differences between groups by severity of COVID-19 related to the previous administration of steroids. However, it is important to note that steroids were primarily administered to critically ill patients.
Additionally, we found a moderate negative correlation between the severity of COVID-19 and the genera Lawsonella and Cutibacterium. Interestingly, although the genus Corynebacterium did not exhibit significant differences among groups, it showed a negative correlation with COVID-19 severity. In contrast, we observed a significant positive correlation between the severity of COVID-19 and the presence of oral cavity-associated genera in nasopharyngeal samples.
In this context, we suggest that this invasion of oral microorganisms may result from the disruption of physical-chemical barriers caused by inflammation during COVID-19, leading to ecological niche competition. Particularly, critically ill COVID-19 patients experience increased mouth breathing before mechanical ventilation support, facilitating the entry of microorganisms such as Streptococcus, Rothia, Veillonella, and Granulicatella from the oral cavity into the respiratory tract. Moreover, intubation disrupts barriers, allowing the invasion of microorganisms such as Prevotella_7, Actinomyces, Solobacterium, and Atopobium into the nasopharynx.
Some of these bacterial genera are considered part of the healthy human microbiome and are found in various anatomical sites [38], primarily in the oral microbiome [39]. These bacteria have also been linked to oral diseases and pathologies in other organs or systems when there is ectopic colonization [[39], [40], [41]]. In the case of the Streptococcus genus in the oral cavity, its presence has been documented on the tongue, mucous membranes, saliva, and dentogingival plaque. Interestingly, different species colonize distinct parts of the mouth [39]. Importantly, Streptococcus pneumoniae has been associated with high case-fatality rates in COVID-19 patients [16].
Rothia is primarily found on the dorsum of the tongue and is associated with nitrate reduction in the oral cavity [39]. However, its translocation to other parts of the body can lead to endocarditis or pneumonia [42]. Veillonella is present in almost the entire oral cavity of healthy individuals [39]. Additionally, an increase in its abundance has been observed in patients with caries [43] and has been reported as a cause of lung abscesses with empyema [44]. Granulicatella is part of the periodontal microbiota in healthy patients, and its decrease has been described in patients with periodontitis [45]. Nevertheless, its translocation to the bloodstream can lead to endocarditis [46].
Prevotella has been described in various anatomical sites [38], as well as throughout the mouth and saliva of healthy individuals, being the second most abundant genus after Streptococcus [47]. It is associated with both periodontal disease and endodontic infections [47] and has been reported as a cause of aspiration pneumonia [48] and Lemierre syndrome characterized by vein thrombosis, oropharyngeal infection, and metastatic septic emboli [49]. Furthermore, in the murine model, Prevotella intermedia has a synergistic effect on pneumococcal pneumonia, increasing inflammatory cytokine levels, bacterial loads in the lungs, and mortality [50]. Actinomyces is present in subgingival plaque and on the tongue [39] and is associated with dental caries [51]. It is important to mention that Actinomyces israelii is the causative agent of actinomycosis, a chronic suppurative granulomatous infection and, in some cases, can be complicated by presenting microabscesses, pneumonia, or septicemia [52].
The potential clinical implications of our findings suggest that the invasion of microorganisms from the oral cavity initiates before intubation, highlighting the clinical relevance of oral hygiene. Oral hygiene is already recognized as an important approach to reducing the risk of ventilator-associated pneumonia [53]. Sampson et al. [54], suggest that there is a link between poor oral health and COVID-19 complications. We highlighted the need for further research to explore strategies like oral hygiene in severe COVID-19 patients to mitigate the risk of critical illness.
Our analysis of data from other studies involving COVID-19 patients categorized by severity revealed varying taxonomic changes, suggesting potential influences of geographic variables, genetics, and diet. Understanding the stability of the microbiome is crucial for elucidating its role in host health and disease susceptibility. Across multiple studies from different countries, we consistently observed the presence of Staphylococcus, Corynebacterium, and Streptococcus genera in samples from healthy volunteers. While some species within these genera are known to be beneficial, others may have pathogenic effects, raising questions about their functional significance in respiratory health.
Lawsonella and Cutibacterium are commonly found in the nasal cavity [55], facial skin [56], and nasopharynx [57]. Cutibacterium has been reported to promote skin health and produce substances with antimicrobial properties [58], suggesting potential roles as commensals within the respiratory tract. Nevertheless, it has also been reported as a human pathogen [59,60]. Furthermore, it is possible that these bacteria were carried out during sampling via nasopharyngeal swabs. Hence, the presence of some taxonomic groups in the nasopharynx or potential consideration as contaminants from the nasal cavity or skin warrants further investigation.
In other viral respiratory infections, such as influenza, specific dominant taxonomic groups are recognized, often linked with pathobionts or opportunistic pathogens such as Acinetobacter baumannii, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Streptococcus pneumoniae [61,62]. In adults with Rhinovirus infection, Allen et al. observed a decline in Haemophilus and Neisseria, alongside an increase in Propionibacterium [63]. Additionally, in adults with Respiratory Syncytial Virus (RSV) infection, Cuthbertson et al. found no statistically significant differences in taxonomic groups or diversity in oropharyngeal samples [64]. These findings highlight the importance of continuing research to shed light on how each viral infection alters physicochemical conditions, immune responses, the microbiome, and anatomical barriers.
Our study has some limitations including, the small sample size, temporal considerations due to variations in the circulating COVID-19 variants, and limited taxonomic resolution. However, it is important to highlight that recruitment of cases of severe and critically ill COVID-19 was done carefully and using strict inclusion criteria in order to avoid bias in the selection of patients and controls.
In conclusion, our study suggests that inflammatory processes and intubation procedures in critically ill COVID-19 patients may disrupt physicochemical conditions, immune responses, microbiome, and barriers. This disruption could potentially facilitate the colonization and invasion of oral microorganisms in the nasopharynx and may be correlated with the severity of COVID-19.
Funding source
This work was supported by internal resources of Instituto de Biotecnología and Instituto Nacional de Medicina Genómica. DGC was supported by a scholarship 2019-000002-01NACF-12598 from Consejo Nacional de Humanidades Ciencias y Tecnología, México.
Ethical approval statement
The study was approved by the Ethics Committee of the INER from Mexico (approval number No.0352-2150). Informed consent was obtained from all subjects.
Data availability statement
The raw V4 region of 16S rRNA sequencing data of this study has been deposited in the NCBI Bioproject database under accession number PRJNA981220. The R script used for this analysis has been uploaded on https://github.com/David-microbiomics/Rscript/blob/main/Rscript_16SV4.
CRediT authorship contribution statement
David Galeana-Cadena: Writing – review & editing, Writing – original draft, Visualization, Validation, Supervision, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. Gustavo Ramirez-Martínez: Writing – original draft, Supervision, Methodology, Formal analysis. José Alberto Choreño-Parra: Supervision, Formal analysis, Conceptualization. Eugenia Silva-Herzog: Writing – original draft, Supervision, Conceptualization. Carmen Margarita Hernández-Cárdenas: Writing – original draft, Supervision, Conceptualization. Xavier Soberón: Writing – review & editing, Writing – original draft, Validation, Supervision, Conceptualization. Joaquín Zúñiga: Writing – review & editing, Writing – original draft, Validation, Supervision, Formal analysis, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors would like to express their gratitude for the invaluable assistance provided by the healthcare workers at the INER, as well as the sequencing unit at the CIENI led by Sandra Maria Pinto Cardoso. We would also like to acknowledge Carlos Federico Arias Ortiz, Pavel Isa, and Humberto Flores Soto for their support and training. Our thanks go to Jerome Verleyen for his technical assistance and for granting us access to the HPC infrastructure at the Unidad Universitaria de Secuenciación Masiva y Bioinformática, Instituto de Biotecnología (UNAM), which is part of the Laboratorio Nacional de Apoyo Tecnológico a las Ciencias Genómicas (CONACyT). Lastly, we would like to thank the Instituto de Biotecnología-UNAM for granting us access to their computer cluster.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e31562.
Contributor Information
Xavier Soberón, Email: xavier.soberon@ibt.unam.mx.
Joaquín Zúñiga, Email: joaquin.zuniga@iner.gob.mx.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Flynn M., Lyall Z., Shepherd G., Lee O.N.Y., Marianna Da Fonseca I., Dong Y., Chalmers S., Hare J., Thomson J., Millar F. Interactions of the bacteriome, virome, and immune system in the nose. FEMS Microbes. 2022;3:1–6. doi: 10.1093/femsmc/xtac020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hardy B.L., Merrell D.S. Friend or foe: Interbacterial competition in the nasal cavity. J. Bacteriol. 2020;203 doi: 10.1128/JB.00480-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cleary D.W., Clarke S.C. The nasopharyngeal microbiome. Emerg. Top. Life Sci. 2017;1:297–312. doi: 10.1042/ETLS20170041. [DOI] [PubMed] [Google Scholar]
- 4.Ren L., Wang Y., Zhong J., Li X., Xiao Y., Li J., Yang J., Fan G., Guo L., Shen Z., Kang L., Shi L., Li Q., Li J., Di L., Li H., Wang C., Wang Y., Wang X., Zou X., Rao J., Zhang L., Wang J., Huang Y., Cao B., Wang J., Li M. Dynamics of the upper respiratory tract microbiota and its association with mortality in COVID-19. Am. J. Respir. Crit. Care Med. 2021;204:1379–1390. doi: 10.1164/rccm.202103-0814OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bacali C., Vulturar R., Buduru S., Cozma A., Fodor A., Chis A., Lucaciu O., Damian L., Moldovan M.L. Oral microbiome : Getting to know and befriend neighbors , a biological approach. Biomedicines. 2022:1–22. doi: 10.3390/biomedicines10030671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prosser J.I., Bohannan B.J.M., Curtis T.P., Ellis R.J., Firestone M.K., Freckleton R.P., Green J.L., Green L.E., Killham K., Lennon J.J., Osborn A.M., Solan M., van der Gast C.J., Young J.P.W. The role of ecological theory. Nat. Rev. Microbiol. 2007;5:384–392. doi: 10.1038/nrmicro1643. [DOI] [PubMed] [Google Scholar]
- 7.Quinn R.A., Comstock W., Zhang T., Morton J.T., Da Silva R., Tran A., Aksenov A., Nothias L.F., Wangpraseurt D., Melnik A.V., Ackermann G., Conrad D., Klapper I., Knight R., Dorrestein P.C. Niche partitioning of a pathogenic microbiome driven by chemical gradients. Sci. Adv. 2018;4 doi: 10.1126/sciadv.aau1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bale R., Iida A., Yamakawa M., Li C.G., Tsubokura M. Quantifying the COVID19 infection risk due to droplet/aerosol inhalation. Sci. Rep. 2022;12:1–15. doi: 10.1038/s41598-022-14862-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Menni C., Valdes A.M., Polidori L., Antonelli M., Penamakuri S., Nogal A., Louca P., May A., Figueiredo J.C., Hu C., Molteni E., Canas L., Österdahl M.F., Modat M., Sudre C.H., Fox B., Hammers A., Wolf J., Capdevila J., Chan A.T., David S.P., Steves C.J., Ourselin S., Spector T.D. Symptom prevalence, duration, and risk of hospital admission in individuals infected with SARS-CoV-2 during periods of omicron and delta variant dominance: a prospective observational study from the ZOE COVID Study. Lancet (London, England) 2022;399:1618–1624. doi: 10.1016/S0140-6736(22)00327-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ball L., Silva P.L., Giacobbe D.R., Bassetti M., Zubieta-Calleja G.R., Rocco P.R.M., Pelosi P. Understanding the pathophysiology of typical acute respiratory distress syndrome and severe COVID-19. Expert Rev. Respir. Med. 2022;16:437–446. doi: 10.1080/17476348.2022.2057300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharma N.K., Sarode S.C. Low pH and temperature of airway surface liquid are key determinants that potentiate SARS-CoV-2 infectivity. Curr. Mol. Med. 2021;22:471–477. doi: 10.2174/1566524021666210816095557. [DOI] [PubMed] [Google Scholar]
- 12.Capelle C.M., Ciré S., Domingues O., Ernens I., Hedin F., Fischer A., Snoeck C.J., Ammerlaan W., Konstantinou M., Grzyb K., Skupin A., Carty C.L., Hilger C., Gilson G., Celebic A., Wilmes P., Del Sol A., Kaplan I.M., Betsou F., Abdelrahman T., Cosma A., Vaillant M., Fagherazzi G., Ollert M., Hefeng F.Q. Combinatorial analysis reveals highly coordinated early-stage immune reactions that predict later antiviral immunity in mild COVID-19 patients. Cell Reports Med. 2022;3 doi: 10.1016/j.xcrm.2022.100600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oliva A., Miele M.C., Di Timoteo F., De Angelis M., Mauro V., Aronica R., Al Ismail D., Ceccarelli G., Pinacchio C., d'Ettorre G., Mascellino M.T., Mastroianni C.M. Persistent systemic microbial translocation and intestinal damage during coronavirus disease-19. Front. Immunol. 2021;12:1–9. doi: 10.3389/fimmu.2021.708149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma S., Zhang F., Zhou F., Li H., Ge W., Gan R., Nie H., Li B., Wang Y., Wu M., Li D., Wang D., Wang Z., You Y., Huang Z. Metagenomic analysis reveals oropharyngeal microbiota alterations in patients with COVID-19. Signal Transduct. Target. Ther. 2021;6 doi: 10.1038/s41392-021-00614-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rattanaburi S., Sawaswong V., Chitcharoen S., Sivapornnukul P., Nimsamer P., Suntronwong N., Puenpa J., Poovorawan Y., Payungporn S. Bacterial microbiota in upper respiratory tract of COVID-19 and influenza patients. Exp. Biol. Med. 2022;247:409–415. doi: 10.1177/15353702211057473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amin-Chowdhury Z., Aiano F., Mensah A., Sheppard C.L., Litt D., Fry N.K., Andrews N., Ramsay M.E., Ladhani S.N. Impact of the coronavirus disease 2019 (COVID-19) pandemic on invasive pneumococcal disease and risk of pneumococcal coinfection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): prospective national cohort study, england. Clin. Infect. Dis. 2021;72:E65–E75. doi: 10.1093/cid/ciaa1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feldman C., Anderson R. The role of co-infections and secondary infections in patients with COVID-19. Pneumonia. 2021;13 doi: 10.1186/s41479-021-00083-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Braun T., Halevi S., Hadar R., Efroni G., Glick Saar E., Keller N., Amir A., Amit S., Haberman Y. SARS-CoV-2 does not have a strong effect on the nasopharyngeal microbial composition. Sci. Rep. 2021;11:8922. doi: 10.1038/s41598-021-88536-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Engen P.A., Naqib A., Jennings C., Green S.J., Landay A., Keshavarzian A., Voigt R.M. Nasopharyngeal microbiota in SARS-CoV-2 positive and negative patients. Biol. Proced. Online. 2021;23:1–6. doi: 10.1186/s12575-021-00148-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prasad P., Mahapatra S., Mishra R., Murmu K.C., Aggarwal S., Sethi M., Mohapatra P., Ghosh A., Yadav R., Dodia H., Ansari S.A., De S., Singh D., Suryawanshi A., Dash R., Senapati S., Beuria T.K., Chattopadhyay S., Syed G.H., Swain R., Raghav S.K., Parida A. Long-read 16S-seq reveals nasopharynx microbial dysbiosis and enrichment of Mycobacterium and Mycoplasma in COVID-19 patients: a potential source of co-infection. Mol. Omi. 2022;18:490–505. doi: 10.1039/d2mo00044j. [DOI] [PubMed] [Google Scholar]
- 21.Mostafa H.H., Fissel J.A., Fanelli B., Bergman Y., Gniazdowski V., Dadlani M., Carroll K.C., Colwell R.R., Simner P.J. Metagenomic next-generation sequencing of nasopharyngeal specimens collected from confirmed and suspect covid-19 patients. mBio. 2020;11:1–13. doi: 10.1128/mBio.01969-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feehan A.K., Rose R., Nolan D.J., Spitz A.M., Graubics K., Colwell R.R., Garcia-Diaz J., Lamers S.L. Nasopharyngeal microbiome community composition and structure is associated with severity of COVID-19 disease and breathing treatment. Appl. Microbiol. 2021;1:177–188. doi: 10.3390/applmicrobiol1020014. [DOI] [Google Scholar]
- 23.Hernández-Terán A., Mejía-Nepomuceno F., Herrera M.T., Barreto O., García E., Castillejos M., Boukadida C., Matias-Florentino M., Rincón-Rubio A., Avila-Rios S., Mújica-Sánchez M., Serna-Muñoz R., Becerril-Vargas E., Guadarrama-Pérez C., Ahumada-Topete V.H., Rodríguez-Llamazares S., Martínez-Orozco J.A., Salas-Hernández J., Pérez-Padilla R., Vázquez-Pérez J.A. Dysbiosis and structural disruption of the respiratory microbiota in COVID-19 patients with severe and fatal outcomes. Sci. Rep. 2021;11 doi: 10.1038/s41598-021-00851-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Merenstein C., Liang G., Whiteside S.A., Cobián-Güemes A.G., Merlino M.S., Taylor L.J., Glascock A., Bittinger K., Tanes C., Graham-Wooten J., Khatib L.A., Fitzgerald A.S., Reddy S., Baxter A.E., Giles J.R., Oldridge D.A., Meyer N.J., Wherry E.J., McGinniss J.E., Bushman F.D., Collman R.G. Signatures of COVID-19 severity and immune response in the respiratory tract microbiome. mBio. 2022;13 doi: 10.1128/mbio.02293-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith N., Goncalves P., Charbit B., Grzelak L., Beretta M., Planchais C., Bruel T., Rouilly V., Bondet V., Hadjadj J., Yatim N., Pere H., Merkling S.H., Ghozlane A., Kernéis S., Rieux-Laucat F., Terrier B., Schwartz O., Mouquet H., Duffy D., Di Santo J.P. Distinct systemic and mucosal immune responses during acute SARS-CoV-2 infection. Nat. Immunol. 2021;22:1428–1439. doi: 10.1038/s41590-021-01028-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.López-Filloy M., Cortez F.J., Gheit T., Cruz y Cruz O., Cruz-Talonia F., Chávez-Torres M., Arteaga-Gómez C., Mancilla-Herrera I., Montesinos J.J., Cortés-Morales V.A., Aguilar C., Tommasino M., Pinto-Cardoso S., Rocha-Zavaleta L. Altered vaginal microbiota composition correlates with human papillomavirus and mucosal immune responses in women with symptomatic cervical ectopy. Front. Cell. Infect. Microbiol. 2022;12:1–15. doi: 10.3389/fcimb.2022.884272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Callahan B.J., McMurdie P.J., Rosen M.J., Han A.W., Johnson A.J.A., Holmes S.P. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar M.S., Slud E.V., Okrah K., Hicks S.C., Hannenhalli S., Corrada Bravo H. Analysis and correction of compositional bias in sparse sequencing count data. BMC Genom. 2018;19:1–23. doi: 10.1186/s12864-018-5160-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lahti S., L, Shetty Microbiome R package. Bioconductor. 2017 doi: 10.18129/B9.bioc.microbiome. [DOI] [Google Scholar]
- 30.Patil I. Visualizations with statistical details: the “ggstatsplot” approach. J. Open Source Softw. 2021;6:3167. doi: 10.21105/joss.03167. [DOI] [Google Scholar]
- 31.Sjoberg D.D., Whiting K., Curry M., Lavery J.A., Larmarange J. Reproducible summary tables with the gtsummary package. R J. 2021;13:570–580. doi: 10.32614/rj-2021-053. [DOI] [Google Scholar]
- 32.Hurst J.H., McCumber A.W., Aquino J.N., Rodriguez J., Heston S.M., Lugo D.J., Rotta A.T., Turner N.A., Pfeiffer T.S., Gurley T.C., Moody M.A., Denny T.N., Rawls J.F., Clark J.S., Woods C.W., Kelly M.S. Age-related changes in the nasopharyngeal microbiome are associated with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection and symptoms among children, adolescents, and young adults. Clin. Infect. Dis. 2022;75:E928–E937. doi: 10.1093/cid/ciac184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kelly B.J., Imai I., Bittinger K., Laughlin A., Fuchs B.D., Bushman F.D., Collman R.G. Composition and dynamics of the respiratory tract microbiome in intubated patients. Microbiome. 2016;4:1–13. doi: 10.1186/s40168-016-0151-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Merenstein C., Bushman F.D., Collman R.G. Alterations in the respiratory tract microbiome in COVID-19: current observations and potential significance. Microbiome. 2022;10:1–14. doi: 10.1186/s40168-022-01342-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shilts M.H., Rosas-Salazar C., Strickland B.A., Kimura K.S., Asad M., Sehanobish E., Freeman M.H., Wessinger B.C., Gupta V., Brown H.M., Boone H.H., Patel V., Barbi M., Bottalico D., O'Neill M., Akbar N., Rajagopala S.V., Mallal S., Phillips E., Turner J.H., Jerschow E., Das S.R. Severe COVID-19 is associated with an altered upper respiratory tract microbiome. Front. Cell. Infect. Microbiol. 2022;11:1–11. doi: 10.3389/fcimb.2021.781968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ventero M.P., Moreno-Perez O., Molina-Pardines C., Paytuví-Gallart A., Boix V., Escribano I., Galan I., González-delaAleja P., López-Pérez M., Sánchez-Martínez R., Merino E., Rodríguez J.C. Nasopharyngeal Microbiota as an early severity biomarker in COVID-19 hospitalised patients. J. Infect. 2022;84:329–336. doi: 10.1016/j.jinf.2021.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reuben R.C., Beugnon R., Jurburg S.D. COVID-19 alters human microbiomes: a meta-analysis. Front. Cell. Infect. Microbiol. 2023;13:1–10. doi: 10.3389/fcimb.2023.1211348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lloyd-Price J., Mahurkar A., Rahnavard G., Crabtree J., Orvis J., Hall A.B., Brady A., Creasy H.H., McCracken C., Giglio M.G., McDonald D., Franzosa E.A., Knight R., White O., Huttenhower C. Strains, functions and dynamics in the expanded human microbiome project. Nature. 2017;550:61–66. doi: 10.1038/nature23889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baker J.L., Mark Welch J.L., Kauffman K.M., McLean J.S., He X. The oral microbiome: diversity, biogeography and human health. Nat. Rev. Microbiol. 2023 doi: 10.1038/s41579-023-00963-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peng X., Cheng L., You Y., Tang C., Ren B., Li Y., Xu X., Zhou X. Oral microbiota in human systematic diseases. Int. J. Oral Sci. 2022;14:1–11. doi: 10.1038/s41368-022-00163-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Willis J.R., Gabaldón T. The human oral microbiome in health and disease: from sequences to ecosystems. Microorganisms. 2020;8:1–28. doi: 10.3390/microorganisms8020308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fatahi-Bafghi M. Characterization of the Rothia spp. and their role in human clinical infections. Infect. Genet. Evol. 2021;93 doi: 10.1016/j.meegid.2021.104877. [DOI] [PubMed] [Google Scholar]
- 43.Teixeira N.D., Almeida de Lima A.K., Do T., Stefani C.M. Meta-analysis using NGS data: the Veillonella species in dental caries. Front. Oral Heal. 2021;2 doi: 10.3389/froh.2021.770917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martinez K., Mangat G.K., Sherwani N., Glover Do M., Silver Md M. Veillonella intrapulmonary abscess with empyema. Cureus. 2023;15:1–8. doi: 10.7759/cureus.45210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu B., Faller L.L., Klitgord N., Mazumdar V., Ghodsi M., Sommer D.D., Gibbons T.R., Treangen T.J., Chang Y.C., Li S., Stine O.C., Hasturk H., Kasif S., Segrè D., Pop M., Amar S. Deep sequencing of the oral microbiome reveals signatures of periodontal disease. PLoS One. 2012;7 doi: 10.1371/journal.pone.0037919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Albano C., Bagarello S., Giordano S., Sanfilippo M.F., Comparato C., Scardino G., Garbo V., Boncori G., Condemi A., Cascio A., Colomba C. Granulicatella spp., a causative agent of infective endocarditis in children. Pathogens. 2022;11:1–9. doi: 10.3390/pathogens11121431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tett A., Pasolli E., Masetti G., Ercolini D., Segata N. Prevotella diversity, niches and interactions with the human host. Nat. Rev. Microbiol. 2021;19:585–599. doi: 10.1038/s41579-021-00559-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang X., Xu C., Yang Z., Zhang T., Du F., Zhou X., Wang Y. Aspiration pneumonia caused by Prevotella causing prothorax after pulmonary puncture: a case report. Clin. Lab. 2023;69 doi: 10.7754/Clin.Lab.2023.230605. [DOI] [PubMed] [Google Scholar]
- 49.Sharma A., Mahajan Z., Mehta S., Puri A. Lemierre syndrome: a diagnostic dilemma of critical care in the post-COVID era. BMJ Case Rep. 2024;17 doi: 10.1136/bcr-2023-257143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nagaoka K., Yanagihara K., Morinaga Y., Nakamura S., Harada T., Hasegawa H., Izumikawa K., Ishimatsu Y., Kakeya H., Nishimura M., Kohnoa S. Prevotella intermedia induces severe bacteremic pneumococcal pneumonia in mice with upregulated platelet-activating factor receptor expression. Infect. Immun. 2014;82:587–593. doi: 10.1128/IAI.00943-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tanner A.C.R., Kressirer C.A., Rothmiller S., Johansson I., Chalmers N.I. The caries microbiome: implications for reversing dysbiosis. Adv. Dent. Res. 2018;29:78–85. doi: 10.1177/0022034517736496. [DOI] [PubMed] [Google Scholar]
- 52.Könönen E., Wade W.G. Actinomyces and related organisms in human infections. Clin. Microbiol. Rev. 2015;28:419–442. doi: 10.1128/CMR.00100-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao T., Wu X., Zhang Q., Li C., Hv W., Hua F., Zhao T., Wu X., Zhang Q., Li C., Hv W., Hua F., Zhao T., Wu X., Zhang Q., Li C., Worthington H., Hua F. Oral hygiene care for critically ill patients to prevent ventilator-associated pneumonia. Cochrane Database Syst. Rev. 2020;12(12):1–139. doi: 10.1002/14651858.CD008367.pub4.www.cochranelibrary.com. John Wiley and Sons Ltd. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sampson V., Kamona N., Sampson A. Could there be a link between oral hygiene and the severity of SARS-CoV-2 infections? Br. Dent. J. 2020;228:971–975. doi: 10.1038/s41415-020-1747-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Natalini J.G., Singh S., Segal L.N. The dynamic lung microbiome in health and disease. Nat. Rev. Microbiol. 2023;21:222–235. doi: 10.1038/s41579-022-00821-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim J.H., Son S.M., Park H., Kim B.K., Choi I.S., Kim H., Huh C.S. Taxonomic profiling of skin microbiome and correlation with clinical skin parameters in healthy Koreans. Sci. Rep. 2021;11:1–15. doi: 10.1038/s41598-021-95734-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yasir M., Al-Sharif H.A., Al-Subhi T., Sindi A.A., Bokhary D.H., El-Daly M.M., Alosaimi B., Hamed M.E., Karim A.M., Hassan A.M., AlShawdari M.M., Alawi M., El-Kafrawy S.A., Azhar E.I. Analysis of the nasopharyngeal microbiome and respiratory pathogens in COVID-19 patients from Saudi Arabia. J. Infect. Public Health. 2023;16:680–688. doi: 10.1016/j.jiph.2023.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rozas M., de Ruijter A.H., Fabrega M.J., Zorgani A., Guell M., Paetzold B., Brillet F. From dysbiosis to healthy skin: major contributions of cutibacterium acnes to skin homeostasis. Microorganisms. 2021;9:1–18. doi: 10.3390/microorganisms9030628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ramesh R., Assi M., Esquer Garrigos Z., Sohail M.R. Lawsonella clevelandensis: an emerging cause of vascular graft infection. BMJ Case Rep. 2021;14 doi: 10.1136/bcr-2020-237350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Prinz J., Schmid B., Zbinden R., Zingg P.O., Uçkay I., Achermann Y., Bosshard P.P. Fast and sensitive multiplex real-time quantitative PCR to detect cutibacterium periprosthetic joint infections. J. Mol. Diagnostics. 2022;24:666–673. doi: 10.1016/j.jmoldx.2022.03.003. [DOI] [PubMed] [Google Scholar]
- 61.Kaul D., Rathnasinghe R., Ferres M., Tan G.S., Barrera A., Pickett B.E., Methe B.A., Das S., Budnik I., Halpin R.A., Wentworth D., Schmolke M., Mena I., Albrecht R.A., Singh I., Nelson K.E., García-Sastre A., Dupont C.L., Medina R.A. Microbiome disturbance and resilience dynamics of the upper respiratory tract during influenza A virus infection. Nat. Commun. 2020;11 doi: 10.1038/s41467-020-16429-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Qin T., Geng T., Zhou H., Han Y., Ren H., Qiu Z., Nie X., Du T., Liang J., Du P., Jiang W., Li T., Xu J. Super-dominant pathobiontic bacteria in the nasopharyngeal microbiota as causative agents of secondary bacterial infection in influenza patients. Emerg. Microbes Infect. 2020;9:605–615. doi: 10.1080/22221751.2020.1737578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Allen E.K., Koeppel A.F., Hendley J.O., Turner S.D., Winther B., Sale M.M. Characterization of the nasopharyngeal microbiota in health and during rhinovirus challenge. Microbiome. 2014;2:1–11. doi: 10.1186/2049-2618-2-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cuthbertson L., James P., Habibi M.S., Thwaites R.S., Paras A., Chiu C., Openshaw P.J.M., Cookson W.O.C., Moffatt M.F. Resilience of the respiratory microbiome in controlled adult RSV challenge study. Eur. Respir. J. 2022;59 doi: 10.1183/13993003.01932-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw V4 region of 16S rRNA sequencing data of this study has been deposited in the NCBI Bioproject database under accession number PRJNA981220. The R script used for this analysis has been uploaded on https://github.com/David-microbiomics/Rscript/blob/main/Rscript_16SV4.