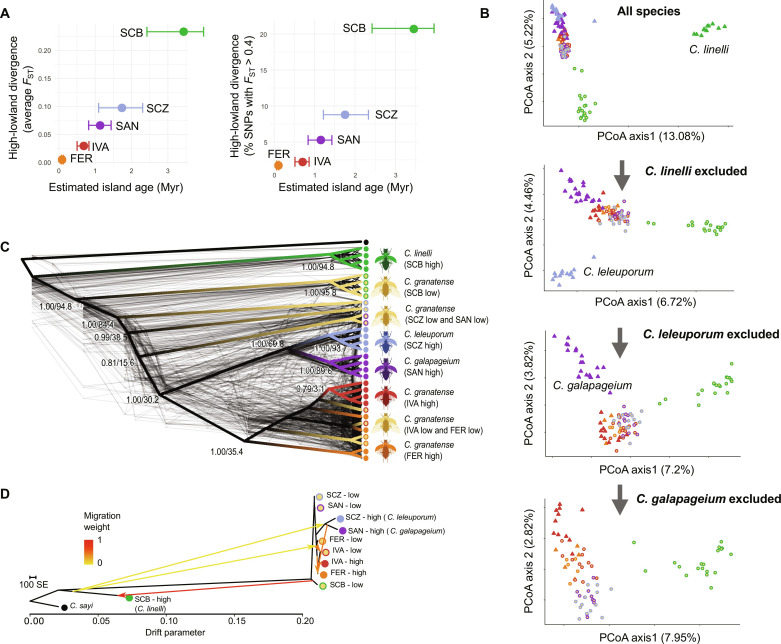
Fig. 3. Genetic differentiation and whole-genome phylogenetic relationships between the Calosoma species and populations from the Galápagos.
(A) Relationship between island age and within-island high-lowland divergence (left panel: average FST; right panel: percentage of SNPs with FST > 0.4) and the estimated age of the island. Estimated ages of each island are based on (36). (B) PCoA of the species and populations based on SNP data obtained from RADtag sequencing. The most divergent highland species was sequentially excluded toward bottom panels. (C) Whole-genome phylogenetic relationship between the species and populations as inferred from 105 ML trees obtained from nonoverlapping genomic windows of 1 Mb (light gray trees), with the multispecies phylogeny [ASTRAL (76)] superimposed. Branch labels show branch support values before slash and gCFs (77), which express the percentage of ML trees containing this branch after the slash. (D) Population tree and population admixture (arrows) of the different Calosoma populations and species, based on SNP data obtained from RADtag sequencing, using TreeMix with six migration edges. Color codes of the different species and populations in all panels are as in Fig. 2. Genomic regions involved in ecotypic high-lowland divergence were excluded for the analyses presented in (B) and (C).