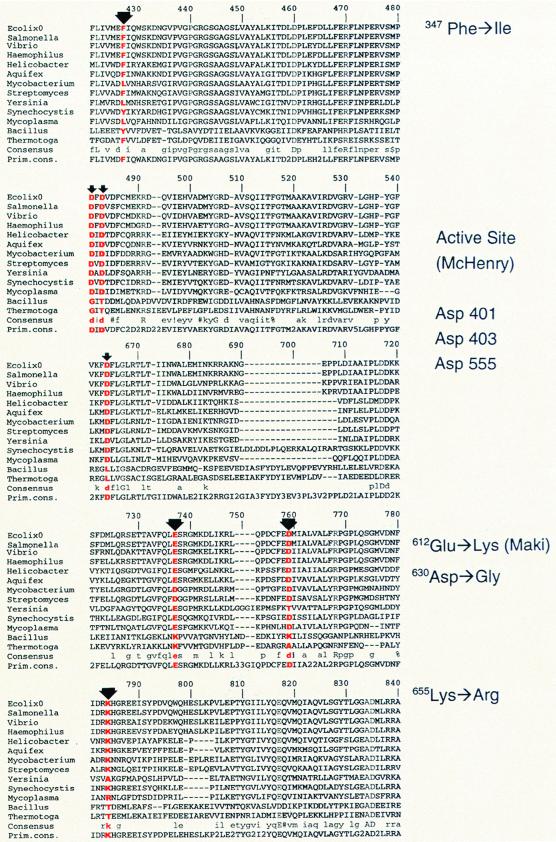
FIG. 1.
Alignment of the E. coli DNA pol III alpha subunit structure with 12 other prokaryotic sequences. Only that portion of the sequence surrounding the sites of the mutator mutants and the active sites (30) is shown. Alignment was done by the Multalin program (4), accessed at http://pbil.ibcp.fr/NPSA. Alignment numbers at the top of the sequence do not refer to E. coli dnaE codons but were assigned by the program to include the gaps required to align all the sequences. The thicker arrows mark the positions of the mutator dnaE alleles used in this study: dnaE1336 (F347I), dnaE1337 (D630G), dnaE1338 (K655R), and dnaE173 (E612K). Thin arrows indicate the position of the active-site aspartic acid residues as defined by Pritchard and McHenry (30). The alleles indicated by the arrows are listed in the margins, with the E. coli dnaE codon numbers given.