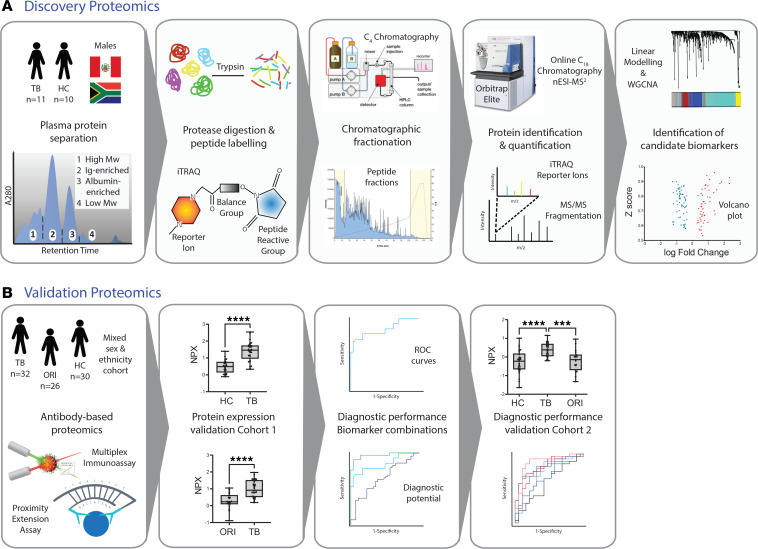
Figure 1. Integrated proteomic study design for TB biomarker identification and validation.
(A) Discovery stage comprising sequential orthogonal fractionation of non-depleted plasma at both the protein and peptide level, iTRAQ peptide labeling, and tandem mass spectrometry for protein identification and relative quantification. Complementary bioinformatic analysis approaches (linear modeling, using limma, and WGCNA) were then used to identify and prioritize diagnostic biomarkers by combining outputs of these pipelines. (B) Candidate protein biomarkers were then validated by multiplex antibody-based techniques (Luminex and proximity extension assay) in serum samples from a separate patient cohort of HCs, pulmonary TB, and ORI of mixed sex and ethnicity. High-performing combinatorial panels were identified for key clinical comparisons and diagnostic performance assessed in 2 separate patient cohorts using binary logistic regression and receiver operating characteristic curves. iTRAQ, isobaric tags for relative and absolute quantification; nESI-MS2, nano-electrospray ionization tandem mass spectrometry; limma, linear modeling for microarray data; WGCNA, whole-gene correlation network analysis; PEA, proximity extension assay; NPX, normalized protein expression; TB, tuberculosis; HC, healthy control; ORI, other respiratory infections; ROC, receiver operating characteristic.