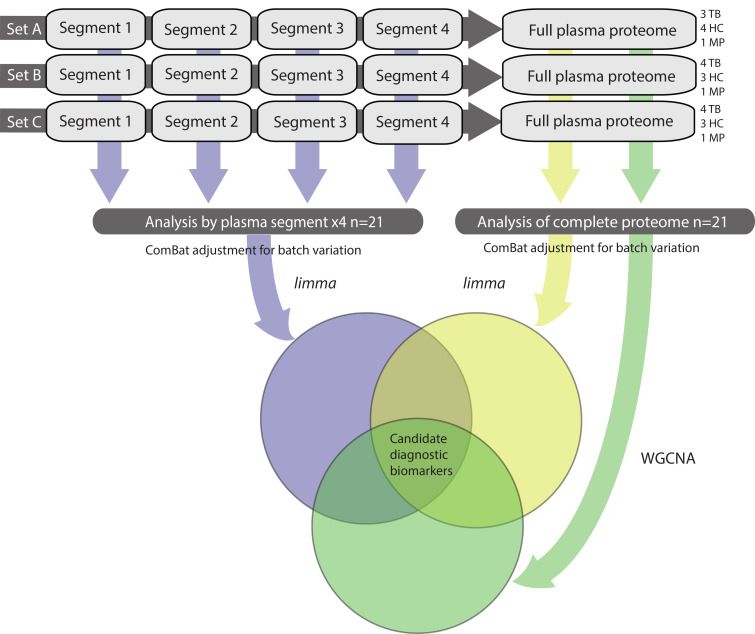
Figure 2. Bioinformatic analysis pipeline.
Discovery proteomics experiments were conducted in 12 separate iTRAQ-labeled 8-plex experiments with block randomization of HC and TB samples into 3 experimental sets. Each plasma segment 8-plex experiment included 1 aliquot of a plasma master pool. Grouped protein abundances were calculated across plasma segments for each experimental set to permit analysis over the whole plasma proteome. Protein abundances were then combined by plasma segment and by experimental set and adjusted for experimental batch variation using ComBat. Differential protein expression was analyzed by limma. In parallel, the complete proteome was analyzed by WGCNA to identify protein networks most strongly correlated with TB. Proteins identified as significant by all 3 bioinformatic approaches were then prioritized for validation. iTRAQ, isobaric tags for relative and absolute quantification; ComBat, adjustment for batch effects using an empirical Bayes framework (R package); WGCNA, whole-gene network correlation analysis; limma, linear modeling for microarray data (R package).