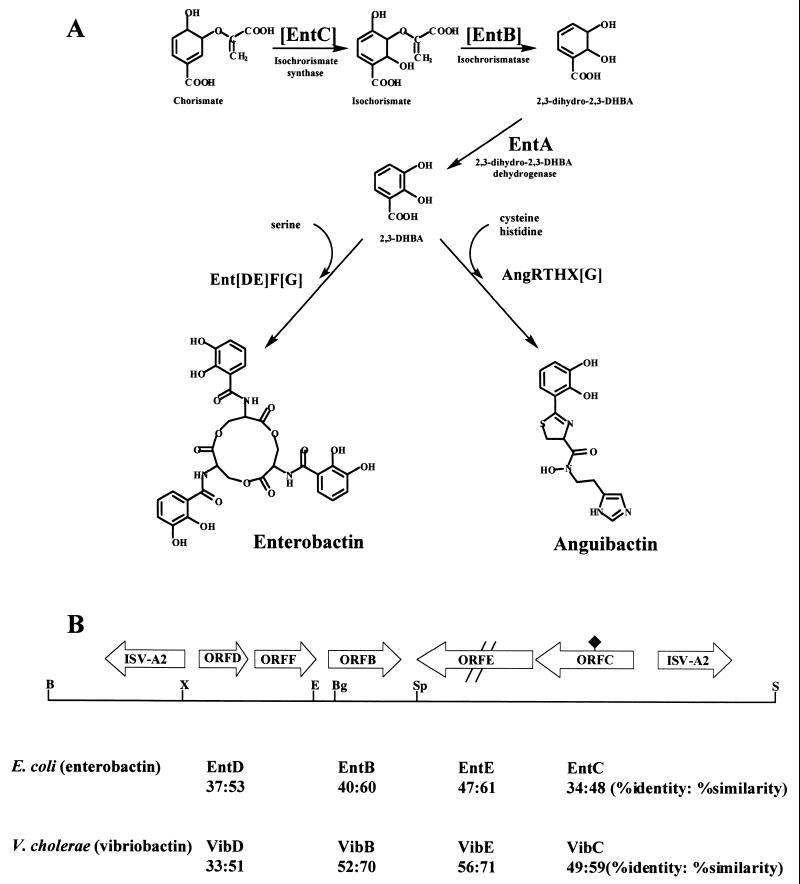
FIG. 1.
Siderophore biosynthesis pathways, physical map of the TAFb region, and homologs of ORFs with siderophore biosynthesis genes. (A) Putative pathways for the biosynthesis of enterobactin and anguibactin. Brackets designate enterobactin biosynthesis enzymes that are homologous to ORFs in the TAFb region. AngR, AngT, and AngH have been characterized previously (45, 52); AngG is characterized in this paper; and X represents other activities required for anguibactin biosynthesis that are still being characterized. (B) Physical map of the locations of ORFs found in the TAFb region and homologies to selected siderophore biosynthesis genes from E. coli and V. cholerae. The double slash symbolizes the frameshift mutation in ORF E; the black diamond corresponds to two translational stop signals in ORF C. B, BamHI; Bg, BglII; E, EcoRV; S, SalI; Sp, SpeI; X, XhoI.