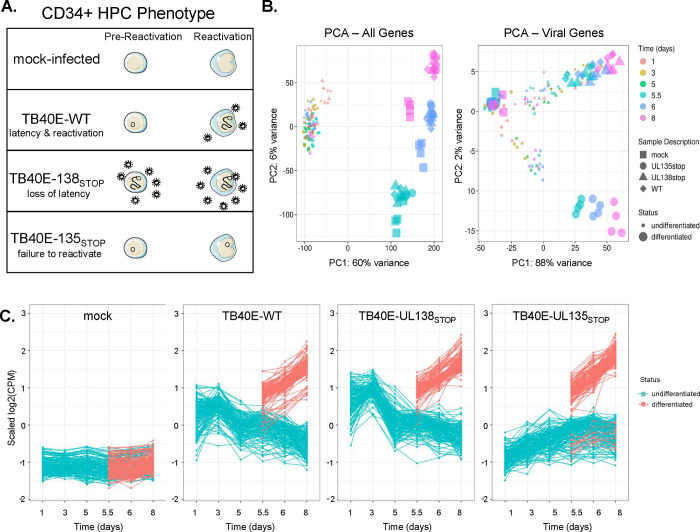
Figure 1. Analysis of the UL135- and UL138-dependent control of the HCMV transcriptome.
A) A depiction of the samples included in this analysis and the phenotype of each virus in our CD34+ HPC model. Mock-infected cells were used to establish a baseline for regulation of cellular genes. Wildtype (WT) virus establishes latency in CD34+ HPCs and reactivates in response to cytokine stimulus. The ΔUL138STOP recombinant is replicative both prior to and following reactivation stimulus (loss of latency) and the ΔUL135STOP recombinant has low replication both prior to and following reactivation stimulus (failure to reactivate). B) Principal Component Analysis (PCA) plots were made using the ggplot2 package (60). Plots were made for both cellular and viral genes (left) and for viral genes only (right). Treatment groups include mock-infected as well as samples infected with WT, ΔUL135STOP, or ΔUL138STOP HCMV. Each treatment group consists of samples collected at 1, 3, and 5 days post infection (dpi) and at 5.5, 6, and 8 dpi treated with either DMSO control or TPA to induce cellular differentiation and viral reactivation. C) A time course of viral gene expression was made for each treatment group. Each set of data points connected by a single line represents one HCMV gene. Data were scaled to log2 counts per million (CPM) as a function of gene count.