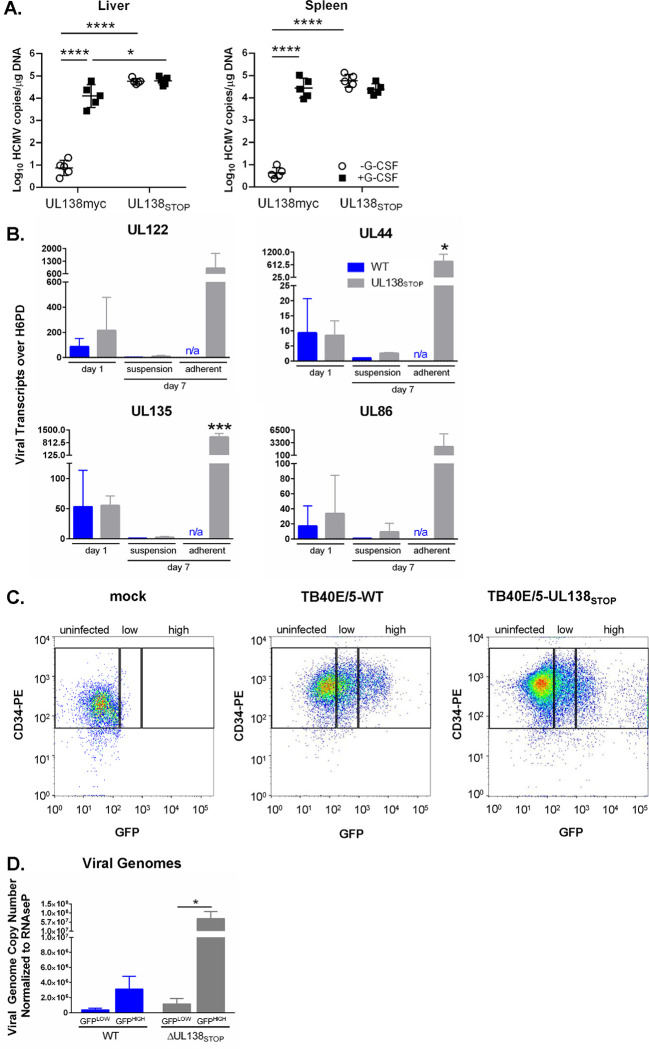
Figure 3. The ΔUL138STOP loss of latency phenotype is pronounced in a subset of HCMV-infected hematopoietic cells.
A) Humanized NSG mice (n = 10 per group) were injected with fibroblasts infected with UL138myc or ΔUL138STOP HCMV. At 4 weeks post infection, half of the mice were treated with G-CSF and AMD-3100 to induce cellular mobilization and trigger viral reactivation. Control mice remained untreated. At 1 week following mobilization, mice were euthanized, and tissues were collected. Total DNA was extracted and HCMV viral load was determined by qPCR using 1 μg of total DNA prepared from liver or spleen tissue. Error bars represent standard error of the mean (SEM) between average vDNA copies from four (liver) or two (spleen) tissue sections for individual animals. All samples were compared by two-way Anova with Tukey’s multiple comparison tests within experimental groups (non-mobilized [−G-CSF] vs mobilized [+G-CSF] for each virus and between all virus groups for both non-mobilized and mobilized conditions). Statistical significance where *, P < 0.05 and ****, P < 0.00005. B) THP-1 cells were infected with WT or ΔUL138STOP HCMV (MOI = 2) and cultured in suspension cell dishes for establishment of latency. Total RNA was extracted at 1 dpi from suspension cells and again at 7 dpi from suspension and adherent cells. cDNA was synthesized and viral transcripts were quantified by RT-qPCR. WT-infected cells did not spontaneously adhere to tissue culture dishes without reactivation stimulus in sufficient quantities to make cDNAs. Error bars represent SEM among three biological replicates analyzed in triplicate. Unpaired t tests were performed to compare individual time points for each virus infection by transcript. Statistical significance where *, P < 0.05 and ***, P < 0.0005. C) CD34+ HPCs were infected with WT or ΔUL138STOP HCMV (MOI = 2) for 24 hours, then CD34/PE+ and GFP+ (infected) cells were isolated by fluorescence-activated cell sorting (FACS). WT- and ΔUL138STOP-infected populations were divided into GFPLOW versus GFPHIGH experimental groups, using the gating strategy shown. D) Pure populations of WT- or ΔUL138STOP-infected CD34+/GFPLOW and CD34+/GFPHIGH cells were cultured over stromal support for establishment of latency. At 10 dpi, total DNA was isolated from each experimental group and viral genomes were quantified by qPCR. Data are shown as viral genome copy number normalized to the cellular gene RNAseP. Three experimental replicates were analyzed in duplicate; error bars represent SEM among experimental replicates.