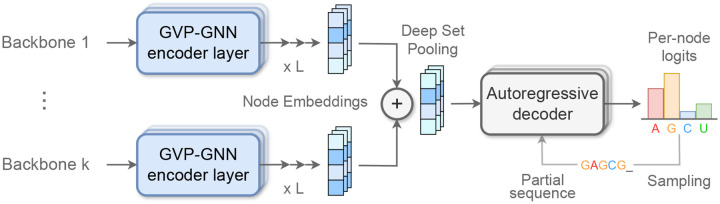
Figure 12: gRNAde model architecture.
One or more RNA backbone geometric graphs are encoded via a series of SE(3)-equivariant Graph Neural Network layers (Jing et al., 2020) to build latent representations of the local 3D geometric neighbourhood of each nucleotide within each state. Representations from multiple states for each nucleotide are then pooled together via permutation invariant Deep Sets (Zaheer et al., 2017), and fed to an autoregressive decoder to predict a probabilities over the four possible bases (A, G, C, U). The probability distribution can be sampled to design a set of candidate sequences. During training, the model is trained end-to-end by minimising a cross-entropy loss between the predicted probability distribution and the true sequence identity.