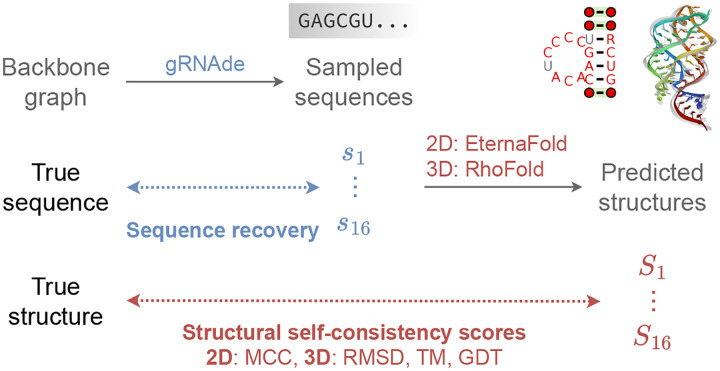
Figure 3: In-silico evaluation metrics for gRNAde designed sequences.
We consider (1) sequence recovery, the percentage of native nucleotides recovered in designed samples, (2) self-consistency scores, which are measured by ‘forward folding’ designed sequences using a structure predictor and measuring how well 2D and 3D structure are recovered (we use EternaFold and RhoFold for 2D/3D structure prediction, respectively). We also report (3) perplexity, the model’s estimate of the likelihood of a sequence given a backbone.