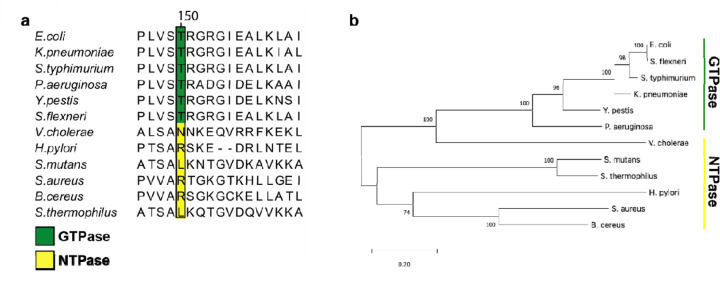
Fig. 5.
Partial multiple sequence alignment (MSA) and phylogenetic analysis of nucleotide-specific and nucleotide-promiscuous FeoBs. a. The partial MSA compares residues in the variable G5 region of experimentally determined GTPases (green) and experimentally determined NTPases (yellow). At position 150 in V. cholerae FeoB (NTPase) is an Asn residue, while the analogous position 150 in the strict GTPase FeoBs is a Thr residue. The numbering above the MSA is based on the V. cholerae FeoB sequence. b. Phylogenetic analysis of the FeoB sequences from the respective organisms in panel ‘a’ shows distinct clustering of the NTPase FeoBs from those of the GTPase FeoBs. The 0.20 scalebar indicates the amount of genetic change at a certain length. The numbers above the nodes represent the bootstrap values above 50% from 500 iterations.