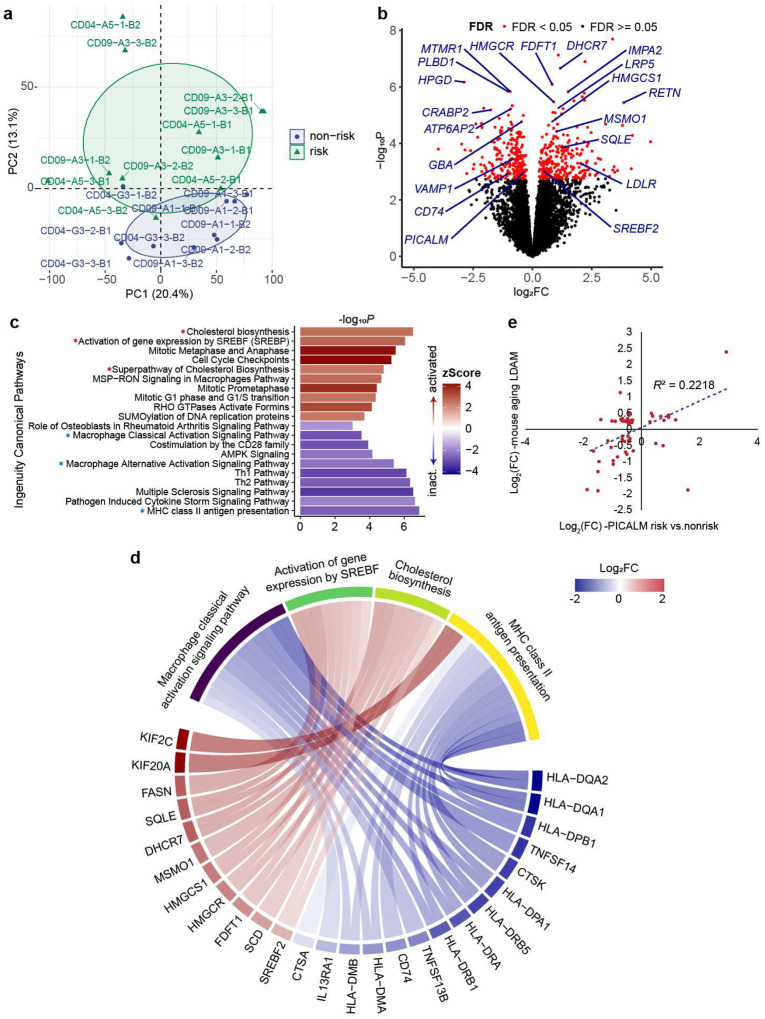
Fig. 4.
Transcriptomic effects of the PICALM LOAD risk allele in iMG. (a) PCA of RNA-seq samples of iMG derived from the isogenic pairs of CRISPR-engineered iPSC lines carrying the PICALM risk or non-risk alleles. Samples are from 2–3 independent cultures of each isogenic pair from 2 different batches for CD04 and CD09 lines. Expression of 13,947 genes was used for PCA. (b) Volcano plot shows DE genes in iMG carrying the LOAD risk allele. (c) Enriched Ingenuity canonical pathways for all DE genes (FDR<0.05). Significantly (FDR<0.05) enriched pathways are ranked by their activated or inactivated Z-scores. (d) Circos plot of DE genes and their expression fold-changes (FC, log2 scale) for major gene pathways highlighted in (c). (e) Significant correlation of the expression changes (-log2FC) in iMG carrying PICALM risk allele (vs. non-risk allele) and in previously reported LD accumulated microglia (LDAM) of the ageing mouse20. Plotted are 56 genes showing DE (FDR<0.1) in both RNA-seq datasets.