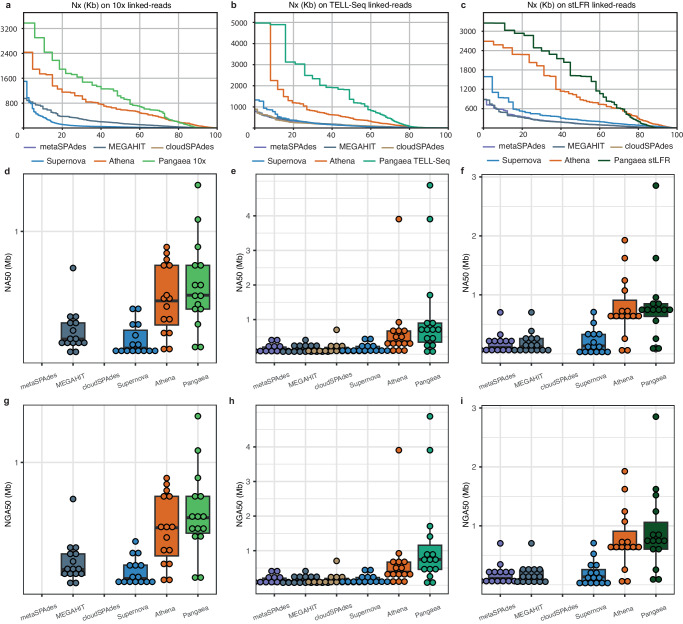
Fig. 2. Contig continuity of assemblies using barcode-removed short-reads (MEGAHIT, metaSPAdes) or linked-reads (cloudSPAdes, Supernova, Athena, Pangaea) from different linked-read sequencing technologies on ATCC-MSA-1003.
Nx, with x ranging from 0 to 100, assembled from 10x Genomics linked-reads (a), TELL-Seq linked-reads (b), and stLFR linked-reads (c) from ATCC-MSA-1003. NA50 for the 15 strains with abundances higher than 0.1% assembled from 10x Genomics linked-reads (d), TELL-Seq linked-reads (e), and stLFR linked-reads (f) on ATCC-MSA-1003. NGA50 for the 15 strains with abundances higher than 0.1% assembled from 10x Genomics linked-reads (g), TELL-Seq linked-reads (h), and stLFR linked-reads (i) on ATCC-MSA-1003. cloudSPAdes was unavailable for 10x Genomics linked-reads and stLFR linked-reads, as it requires extremely large memory (>2TB) on these datasets. The samples are biological replicates for (d–i), n = 15, each stands for a strain with abundance >0.1%. Box plots show the median (center line), 25th percentile (lower bound of box), 75th percentile (upper bound of box), and the minimum and maximum values within 1.5 × IQR (whiskers) as well as outliers (individual points). Source data are provided as a Source Data file.