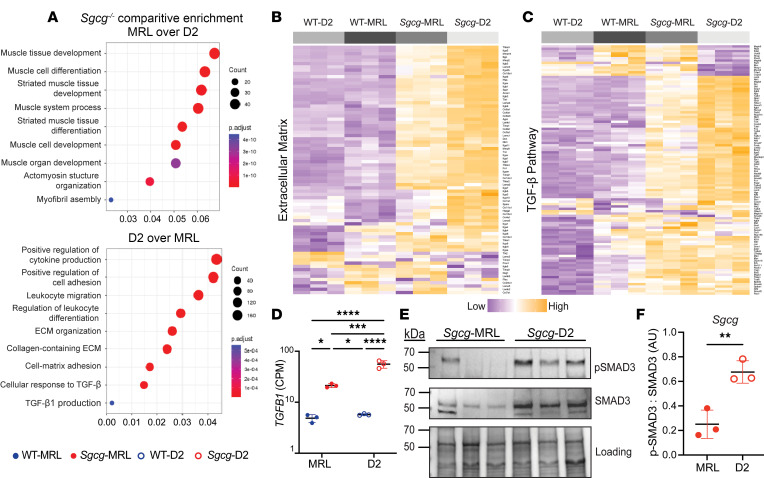
Figure 3. TGF-β gene expression and signaling were downregulated in Sgcg-MRL skeletal muscle compared with Sgcg-D2 muscle.
RNA sequencing was performed on quadriceps muscles from 16-week-old female mice in biological triplicate. Gene expression was normalized to counts per million (CPM). (A) Comparison of Sgcg-MRL versus Sgcg-D2 transcriptomic profiles showed muscle development and differentiation genes to be highly enriched the Sgcg-MRL background (top). In contrast, immune response, extracellular matrix, and TGF-β pathways were highly enriched in the Sgcg-D2 cohort. (B) Clustered heatmap shows reduction of extracellular matrix genes in Sgcg-MRL muscle. (C) Clustered heatmap shows reduction in TGF-β pathway genes in Sgcg-MRL muscle. (D) Comparative log-scale analysis showed substantially higher average expression of Tgfb1 in the Sgcg-D2 cohort (WT-MRL 4.86, Sgcg-MRL 21.3, WT-D2 5.73, and Sgcg-D2 55.9 CPM). (E) Immunoblotting of quadriceps muscles from Sgcg-D2 and Sgcg-MRL mice showed reduced phosphorylated SMAD3 (p-SMAD3) and total SMAD3 in Sgcg-MRL. (F) The ratio of p-SMAD3 to total SMAD3 was quantified and indicated TGF-β signaling was decreased in the MRL background. Data are presented as mean ± SD. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 by 2-way ANOVA (D) or 2-tailed Student’s t test (F).