Figure 2.
Integrative protocol analysis of scRNA-seq data identified synovial structural, myeloid and lymphoid cell populations
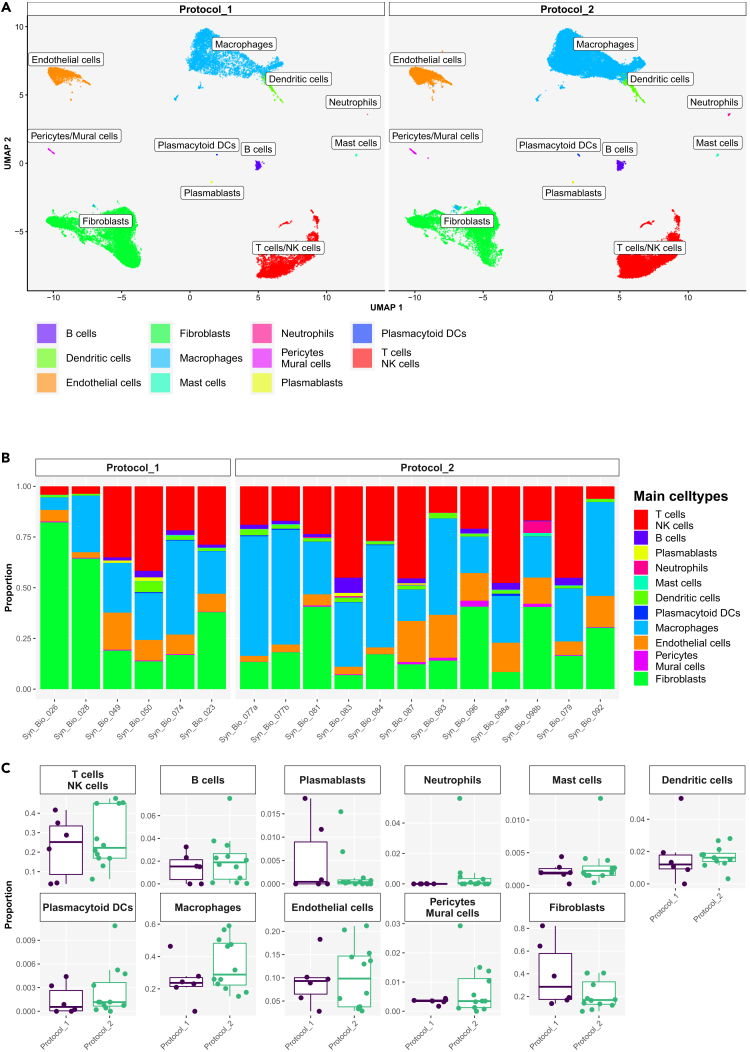
(A) UMAPs of the integrated protocol 1 and protocol 2 scRNA-seq dataset with main synovial cell populations colored by main cell type. ScRNA-seq data generated from fresh human synovia dissociated either with the original protocol of Donlin et al.22 (protocol 1, n = 6) or the optimized protocol (protocol 2, n = 12).
(B) Bar plots of relative abundances of main cell types per sample per protocol. Neutrophils visible primarily in protocol 2 samples, including SynBio_081, SynBio_083 and Syn_Bio_098b.
(C) The proportion of cell types per protocol 1 versus protocol 2; neutrophils were detected primarily in protocol 2 scRNA-seq data, but the difference between the protocols was not statistically significant as calculated by differential abundance analysis in EdgeR. The box plot visualises 5 summary statistics: the median; two hinges, corresponding to the first and the third quartiles; two whiskers. The upper (lower) whisker extends from the hinge to the largest (smallest) value no further than 1.5 ∗ interquartile range from the hinge. Individual dots represent data from different samples.