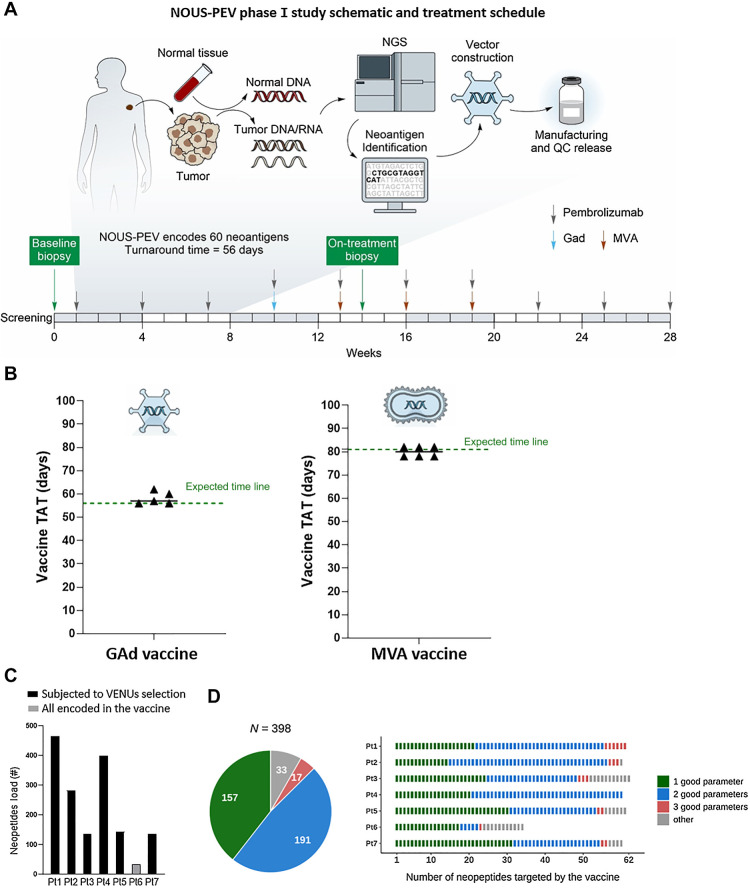
Figure 1.
NOUS-PEV neoantigen vaccines: trial design, feasibility, and selection of encoded neoantigens. A, Schematic outlining the NOUS-PEV trial design, vaccine production, and treatment schedule. B, Achieved (triangles) and expected (green dotted line) turnaround time (TAT) to manufacture and release GAd20 and MVA-PEV. The TAT is calculated in days, from when the biopsy is available for the nucleic acid extraction till the day the vaccine is released. C, Total number of neopeptides detected in NOUS-PEV patients. Each bar represents the total number of neopeptides encoded by nonsynonymous somatic mutations detected in baseline tumor biopsies. Black and gray bars indicate the patients’ mutation subjected or not to VENUS prioritization algorithm, respectively. D, Quality of neopeptides included in NOUS-PEV. The pie chart displays the total number of candidate neopeptides targeted by NOUS-PEV. In green, blue, and salmon are indicated respectively, the selected neopeptides showing one, two, or three of the parameters defined “good” according to the following thresholds (TPM of the RNA carrying the mutation ≥1; MHC class I predicted binding IC50 ≤ 500 nmol/L; mutation allele frequency >50%). In gray are the other neopeptides not included in the previous three categories. The bar plot indicates the detail of candidate neopeptides for each individual patient.