Significance
The accumulation of tau protein aggregates in the brain is one of the hallmarks of Alzheimer’s disease. Tau aggregates are thought to enter cells through the endolysosomal pathway and escape into the cytosol through a poorly understood mechanism. We found that in primary neurons and astrocytes, endocytosis of tau aggregates leads to lysosomal swelling and permeabilization, triggering lysosome deacidification, and recruitment of the membrane-sealing ESCRT machinery. Additionally, new cytosolic tau aggregates appeared on the surface of tau-burdened lysosomes, suggesting that endolysosomal escape of tau aggregates occurs through nanoscale membrane damage.
Keywords: lysosome, Alzheimer’s disease, ESCRT, STORM, astrocyte
Abstract
The prion-like spread of protein aggregates is a leading hypothesis for the propagation of neurofibrillary lesions in the brain, including the spread of tau inclusions associated with Alzheimer’s disease. The mechanisms of cellular uptake of tau seeds and subsequent nucleated polymerization of cytosolic tau are major questions in the field, and the potential for coupling between the entry and nucleation mechanisms has been little explored. We found that in primary astrocytes and neurons, endocytosis of tau seeds leads to their accumulation in lysosomes. This in turn leads to lysosomal swelling, deacidification, and recruitment of ESCRT proteins, but not Galectin-3, to the lysosomal membrane. These observations are consistent with nanoscale damage of the lysosomal membrane. Live cell imaging and STORM superresolution microscopy further show that the nucleation of cytosolic tau occurs primarily at the lysosome membrane under these conditions. These data suggest that tau seeds escape from lysosomes via nanoscale damage rather than wholesale rupture and that nucleation of cytosolic tau commences as soon as tau fibril ends emerge from the lysosomal membrane.
Neurodegenerative diseases of protein misfolding are characterized by the abnormal aggregation of host proteins into amyloid inclusions. These inclusions are defined by the presence of a cross-β structural core (1). The major neurodegenerative diseases, including Alzheimer’s disease (AD), Parkinson’s disease (PD), and amyotrophic lateral sclerosis (ALS), are characterized by progressive spread of these protein aggregates within the brain (2). The leading model for spread is that inclusions propagate by a prion-like mechanism, in which seeds spread from donor cells and nucleate the misfolding and aggregation of endogenous protein in recipient cells (3, 4).
One such class of prion-like neurodegenerative diseases, termed “tauopathies,” is associated with the aggregation of the protein tau. Tau is a microtubule-associated protein encoded by a single gene but alternatively spliced into six expressed isoforms in humans, which vary in the number of N-terminal domains (N) and microtubule-binding repeats (R) (5). Tau aggregation is considered to be a driver of AD and also a larger family of neurodegenerative diseases known as tauopathies (5). Tauopathies are diagnosed by identification of tau protein aggregates in neurons, astrocytes, and oligodendrocytes of postmortem brain tissue (6–9). In general, the structure of tau fibrils is polymorphic, and each tauopathy is characterized by a unique fibril structure as revealed by high-resolution cryo-EM (10, 11).
Tau is thought to spread between cells when seeds are taken up in recipient cells via endocytosis (12). Once seeds have entered recipient cells, tau seeds are capable of nucleating cytosolic tau aggregation (13–15). The pathway and egress of tau seeds from the endocytic and endolysosomal systems have been a major question in the field. One leading hypothesis has been that tau seeds escape via lysosomal rupture. Galectins have been used as a marker for rupture because they bind to the newly exposed lumenal glycans of ruptured lysosomes and trigger their degradation via lysophagy (16). Previous studies conducted in immortalized and primary cells have demonstrated lysosomal accumulation of tau fibrils with evidence of lysosome damage occurring using a Galectin-based reporter system in some reports (17–19), although not in others (14, 15). The inconsistency in these reports suggests that lysosomal rupture, at least on the scale necessary to expose lumenal glycans, cannot be a general explanation for tau escape.
It was recently reported that ESCRT proteins, specifically the ESCRT-III subunit CHMP6, suppressed tau fibril escape from the lysosomal lumen to the cytosol (14). The ESCRT proteins are a conserved machinery for membrane budding and sealing (20, 21) in many pathways in cells, including ectosome and virus release, cytokinetic abscission, multivesicular body biogenesis in the endolysosomal pathway, and membrane repair. Among their many functions, ESCRTs are responsible for repairing nanoscale lysosomal membrane damage (22–25). Damage sensing is thought to be mediated by the leakage of Ca2+ from the lysosome as detected by ALG-2 and other Ca2+-and membrane-binding proteins (22, 23, 26). This form of membrane damage is distinct from the larger ruptures detected by Galectin-3 and triggers a repair response rather than lysophagy (24, 25). Consistent with a potential role for nanoscale lysosomal membrane damage in tau escape, the lysosome membrane damaging agent leucyl-leucyl-O-methyl ester (LLOME) accelerates seeded tau aggregation (14). Nevertheless, ESCRTs act at multiple points in both the biogenesis and repair of lysosomes, and the crucial question of the mechanism of lysosomal escape by tau seeds remains to be determined.
Here, we set out to determine whether tau does in fact escape from lysosomes via nanoscale membrane damage. We found that tau fibril accumulation within human primary astrocyte and neuron lysosomes triggered membrane permeabilization and an increase in lysosomal pH that was sensed by ESCRTs. Using confocal and STORM microscopy, we characterized the ultrastructure of the fibril-burdened lysosomal compartment and identified lysosomes with tau fibrils protruding from their membranes. These findings reveal a direct mechanism where tau seeds induce lysosomal membrane permeabilization and trigger the templated aggregation of cytosolic tau at the surface of the lysosome.
Results
Tau Fibrils Deacidify Lysosomes.
We established a primary human astrocyte-based model of endocytosis of preformed 4R tau fibrils (PFF), which we used to monitor the effect of fibril uptake on the endolysosomal system. In astrocytes, tau accumulation induces the formation of astrocytic plaques and tufted astrocytes, the diagnostic criterion for Corticobasal Degeneration (CBD) and Progressive Supranuclear Palsy (PSP), respectively (8, 9). Tau inclusions in astrocytes are typically composed of a truncated form of tau containing 4 C-terminal repeats (4R tau) (27), which was therefore used to generate fibrils in this study. Astrocytes acquire extracellular tau by a variety of mechanisms (7, 18, 28–31). We therefore carried out the study in astrocytes as a system directly relevant to disease, robust in its response, and tractable for the imaging experiments described below.
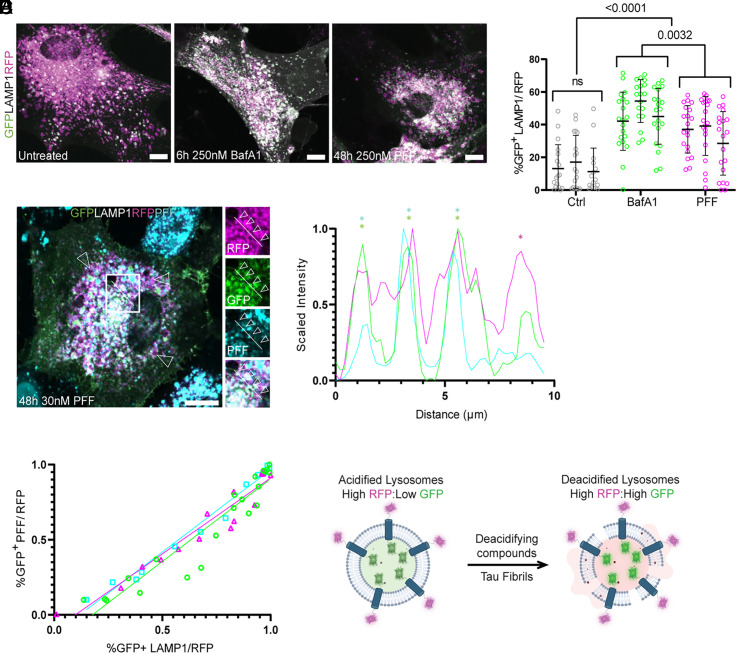
The hypothesis that tau fibrils induced nanoscale damage to the lysosomes predicts that exposure to fibrils should create leaks that increase lysosomal pH. To test for changes in pH, normal human brain-derived astrocytes were transduced with a lysosome activity reporter (pHLARE) which monitors lysosomal pH (Fig. 1A). pHLARE is a late endosome associated protein-1 (LAMP1)-derived construct that is a single-pass transmembrane protein with tandem fusions to lumenal pH-sensitive GFP and cytosolic RFP (32). Healthy lysosomes exhibit a pH of ~5, but loss of a proton gradient due to membrane permeabilization or impairment of the lysosomal proton pumping V-ATPase can drastically raise the pH to ~7, equal to that of the cytosol (32). At rest, approximately 14 ± 15% of astrocyte lysosomes exhibited detectable GFP levels. Treatment with 250 nM Bafilomycin A1 (BafA1) for 6 h, an inhibitor of the V-ATPase, increased the number of lysosomes with high pH to 47 ± 17%. Similarly, although to a lesser extent, incubation with 250 nM PFF for 48 h increased the number of high pH lysosomes to 35 ± 18% (Fig. 1B). Soon after the submission of this manuscript, lysosome deacidification was independently observed in immortalized cells overloaded with tau which revealed an increase in lysosomal pH from 4.76 to 5.147 as measured by a Lysosensor probe (33).
Fig. 1.
Tau fibrils permeabilize lysosomes. Primary human astrocytes were stably transduced with a GFP–LAMP1–RFP tandem fusion protein to stoichiometrically monitor lysosomal pH. At rest, most lysosomes were RFP-positive. After treatment with Bafilomycin or PFF, the fraction of GFP-positive lysosomes increased dramatically (A). Quantification and two-way ANOVA of total cell GFP area normalized to total RFP signal prior to analysis (B) (n = 52, 58, and 60 cells for untreated, BafA1, and PFF conditions, respectively, individual cells from each independent replicate are plotted with mean and SD for each replicate, 15 to 20 cells per each of three replicates). The fibril seeding assay was performed in parallel using 30 nM fluorescently labeled PFF (C). A representative line scan from a cell with four neighboring lysosomes and only one containing labeled fibrils (RFP in magenta, GFP in green, and fibrils in cyan) (D). (E) Line plots revealing a positive correlation between cells that had GFP-positive lysosomes and GFP-positive PFF across three replicates (R2 = 0.89, n = 21 cells; R2 = 0.92, n = 16 cells; R2 = 0.97, n = 13 cells, plotting individual Manders colocalization coefficients). Schematic detailing the effect of deacidifying compounds and tau fibrils on the state of lysosomal pH (F). (Scale bar: 20 µm.)
To determine whether the observed increase in lysosomal pH induced by PFF endocytosis correlated with the presence of PFF in the lysosomal lumen, we repeated our seeding assay with fluorescently labeled PFF. In these experiments, the concentration of PFF added to the media was 30 nM (Fig. 1C). At this lower concentration of tau seeds, we could identify neighboring lysosomes in the same cells that demonstrated clear fibril-dependent pH differences (Fig. 1D). We also detected a strong positive correlation between the lumenal presence of PFF and deacidified lysosomes (Fig. 1E). These data show that burdening the endolysosomal system with PFF potently neutralizes lysosomal pH (Fig. 1F).
ESCRTs, but not Galectins, Are Recruited to PFF-containing Lysosomes.
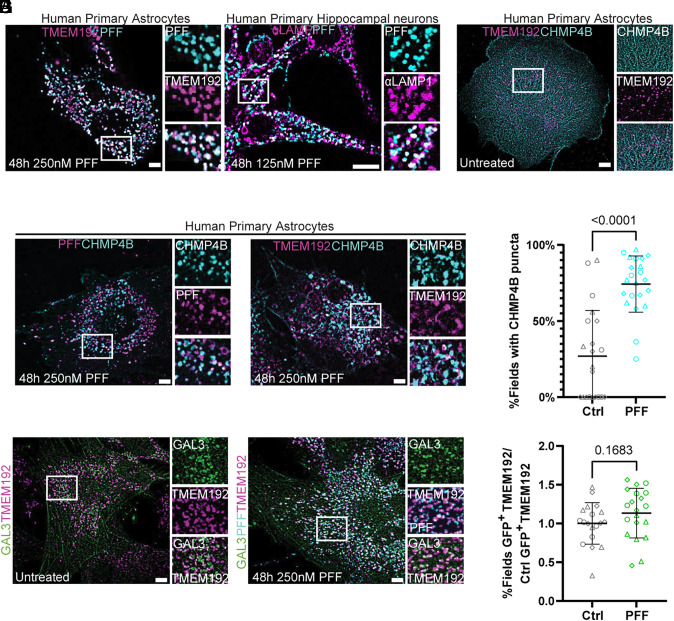
Having observed an effect of PFF on the pH of the lysosomal compartment, we suspected that PFF could be either interfering with V-ATPase function (34, 35), or mechanically damaging lysosomal membranes leading to proton leakage. To test for a lysosome damage response to PFF, we first transduced human primary astrocytes with TMEM192–RFP to label lysosomes and treated cells with fluorescently labeled PFF. PFF accumulated in the lysosomal compartment labeled by TMEM192–RFP, in agreement with previous reports (14, 15) (Fig. 2A). We additionally asked whether the same phenotype was true for human neurons and observed that labeled PFF accumulated in the lysosomes of primary human hippocampal neurons (referred to as neurons hereafter) labeled with endogenous LAMP1 antibody, in agreement with previous reports (36–38) (Fig. 2A). We next transduced TMEM192–RFP expressing astrocytes with CHMP4B, a core component of the ESCRT system (20, 21), fused to a c-terminal HALO tag with a long linker known to support function (39) to test for ESCRT recruitment after PFF treatment. At rest, CHMP4B expression was diffuse with few puncta and did not interact with TMEM192-positive lysosomes (Fig. 2B). After fibril treatment, CHM4B formed large puncta on the surface of lysosomes and PFF, consistent with a triggered nanoscale damage repair response (Fig. 2 C–E). Consistent with other recent reports (14, 15), we did not observe a significant increase in Galectin-3 recruitment to TMEM192–RFP-labeled lysosomes, suggesting that PFF permeabilizes but does not rupture the lysosomes of astrocytes (Fig. 2 F–H). Substantial numbers of Galectin-3 puncta are present in both control and PFF-treated cells, showing that a significant level of lysosomal rupture already exists in the absence of treatment. These data show that in astrocytes, tau fibrils permeabilize but do not rupture lysosomes.
Fig. 2.
CHMP4B, but not Galectin-3, is recruited to fibril-containing lysosomes. TMEM192–RFP expressing astrocytes were treated with labeled PFF to verify endosomal uptake, which was phenocopied by human primary neurons stained with endogenous LAMP1 (A). These same cells were additionally transduced with CHMP4B–HALO to monitor potential ESCRT recruitment to fibril-containing lysosomes. At rest, CHMP4B was mostly diffuse with few puncta that rarely interacted with TMEM192-positive lysosomes (B). Treatment with labeled PFF triggered the robust formation of CHMP4B puncta that overlapped with fibrils and lysosomes (C and D). (E) Quantification of CHMP4B-expressing cells before and after PFF treatment, compared using two-tailed unpaired t test (n = 3 replicates, 20 random and independent fields of 10 to 20 cells per replicate, n = 386 cells, represented as mean and SD across fields). TMEM192–RFP expressing astrocytes were transduced with Gal3–GFP and the fractional overlap of GFP puncta area and endogenous TMEM192–RFP area was compared before and after fibril treatment, compared using two-tailed unpaired t test (F–H) (n = 4, 20 random and independent fields of 10 to 20 cells per replicate, n = 372 cells). (Scale bars: 10 µm.)
Interference with the ESCRT Machinery Worsens Constitutive Lysosome Permeability, Promoting PFF Entry into the Cytosol.
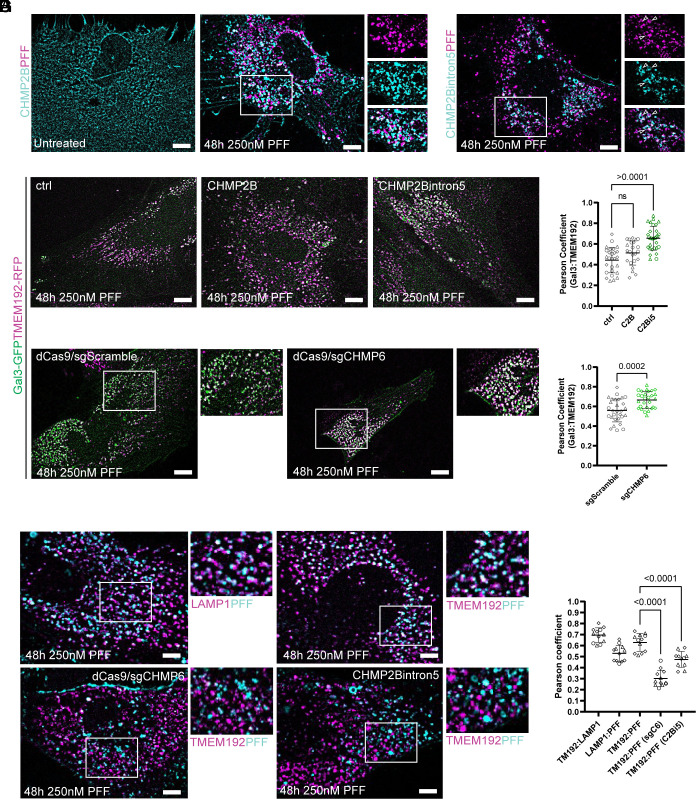
Having observed an induction of the ESCRT membrane repair machinery to damaged lysosomes containing PFF, we next tested the effect of ESCRT perturbation on lysosomal permeability and PFF escape into the cytosol. The membrane scission and sealing function performed by the downstream ESCRT-III proteins requires a recruitment cascade of CHMP proteins including CHMP6 and CHMP2B (20, 21). A familial form of frontotemporal degeneration (FTD) has been linked to a mutation resulting in the expression of a C-terminally truncated CHMP2B, known as CHMP2Bintron5, in patient brains (40, 41). CHMP2Bintron5 is missing its C-terminal microtubule-interacting and transport-interacting motif (MIM) domain which is essential for its removal from membrane scission-competent ESCRT-III polymers by the ESCRT-associated AAA ATPase VPS4 (42). Removal of ESCRT-III polymers is required for successful membrane sealing, and removal of the CHMP2B MIM caused by this mutation blocks ESCRT function, causing membrane trafficking defects and late endosome swelling in patient samples and animal models (40, 43–46). However, it is currently unknown what effect, if any, expression of CHMP2Bintron5 has on the endolysosomal escape of PFF.
More recently, it was demonstrated that the CRISPRi-mediated knockdown of CHMP6 resulted in lysosome permeability and PFF escape in immortalized cells, likely by preventing ESCRT-mediated sealing of lysosomes containing PFF (14). However, the effect of CHMP6 depletion in primary human astrocytes is yet to be tested. We therefore sought to determine the effect of ESCRT inhibition on lysosome permeability and PFF endolysosomal escape using two parallel and complementary approaches of CHMP2Bintron5 overexpression and CHMP6 knockdown.
To first validate the expression and recruitment dynamics of CHMP2Bintron5, primary human astrocytes were stably transduced with CHMP2Bintron5–HALO or CHMP2B–HALO as a control. CHMP2B–HALO phenocopied CHMP4B–HALO and was diffuse at rest but formed puncta and was recruited to PFF after treatment (Fig. 3A). Interestingly, CHMP2Bintron5–HALO was also recruited to PFF but formed enlarged endosomes at rest that also appeared adjacent to PFF signal (Fig. 3B). We next tested the effect of CHMP2Bintron5 overexpression on lysosome permeability. Primary human astrocytes were first stably transduced with TMEM192–RFP and Gal3–GFP as above (Fig. 2). These cells were then additionally transduced with either full-length CHMP2B–HALO or CHMP2Bintron5–HALO and compared to a mock transduced control. All CHMP constructs used in this study have a secondary promoter controlling the constitutive expression of BFP, allowing for the identification of ESCRT-expressing cells without the use of a fluorescent HALO ligand. Since our previous experiments revealed a large fraction of preexisting Gal3-positive lysosomes in untreated cells, we looked for increases in Gal3-positivity due to overexpression of CHMP2Bintron5 in the presence of PFF. Unlike CHMP2B–HALO expression which did not change the Gal3-positivity of lysosomes, CHMP2Bintron5–HALO expression resulted in a significant increase in Gal3-positivity (Fig. 3 C and D). In parallel, Gal3–GFP and TMEM192–RFP-expressing astrocytes were stably transduced with dCas9 BFP to allow for the assessment of lysosome permeability in the context of CRISPRi-mediated ESCRT knockdown (14). These cells were then additionally transduced with scramble sgRNA with a secondary promoter driving BFP expression (sgScramble) or previously validated CHMP6 sgRNA (sgCHMP6) for 3 d prior to a 2-d incubation with PFF. Cells were then assessed for changes in Gal3-lysosome positivity. In agreement with increased Gal3-lysosome positivity from interfering with ESCRT function by CHMP2Bintron5 expression, we also observed a significant increase in Gal3 recruitment to lysosomes after transduction with sgCHMP6 but not sgScramble (Fig. 3 C and E). Therefore, impairment of the ESCRT machinery compromised endolysosome integrity and worsened pre-existing Gal3 positivity.
Fig. 3.
Disruption of the ESCRT machinery exacerbates pre-existing lysosome permeation and enhances PFF escape. TMEM192–RFP expressing astrocytes were stably transduced with CHMP2B–HALO or CHMP2Bintron5–HALO prior to treatment with PFF. (A) CHMP2B–HALO expression was diffuse in untreated cells and puncta formation was induced by PFF. CHMP2Bintron5–HALO puncta were also recruited to PFF, but appeared irregular and often additionally marked enlarged endosomes that were not seen with CHMP2B–HALO or CHMP4B–HALO (B). CHMP2Bintron5 but not CHMP2B expression increased Gal3–GFP lysosome positivity in astrocytes expressing Gal3–GFP and TMEM192–RFP (C). (D) Quantification of Gal3-positive lysosome area represented as mean and SD across replicates, compared by one-way ANOVA (D) (n = 3, 10 independent fields of 10 to 20 cells per replicate). dCas9 was introduced into the Gal3–GFP and TMEM192–RFP dual expressing cells via transduction to test the effect of ESCRT knockdown on lysosome integrity. Cells were then transduced with scramble sgRNA or a previously validated CHMP6 sgRNA for 3 d prior to PFF incubation (C). Expression of sgCHMP6 significantly increased lysosomal galectin positivity compared to the sgScramble control, represented as mean and SD across replicates, compared by two-tailed unpaired t test (E) (n = 3, 10 independent fields of 10 to 20 cells per replicate). (F) TMEM192 and LAMP1 mark the PFF compartment while additional transduction with sgCHMP6 (sgC6) or CHMP2Bintron5–HALO (C2Bi5) promotes PFF translocation to the cytosol. (G) Quantification of PFF signal from the lysosome compartment after ESCRT perturbation as mean and SD across replicates, compared by two-tailed unpaired t test (n = 3, four independent fields of 10 to 20 cells per replicate). (Scale bars: 10 µm.)
Having demonstrated that lysosome permeability was enhanced by interference with the ESCRT machinery, we next investigated whether this impairment correlated with the increased translocation of PFF into the cytosol. Astrocytes expressing Gal3–GFP with TMEM192–RFP additionally transduced with either CHMP2Bintron5 or dCas9/sgCHMP6 were subject to the same assay as in Fig. 3C, but with labeled PFF (Fig. 3F). After 3 d of sgCHMP6 expression or two days with CHMP2Bintron5 overexpression, there was a significant decrease in colocalization of labeled PFF and TMEM192–RFP compared to cells treated with PFF alone, due to increased lysosome permeability (Fig. 3G). We also found that LAMP1–GFP strongly positively correlated with both TMEM192–RFP and PFF, validating its use as a lysosome marker, in agreement with previous reports (14, 15). By interfering with the ESCRT machinery in cells treated with PFF, astrocytes experience a worsening of pre-existing lysosome permeability which promotes the translocation of PFF into the cytosol.
Soluble Tau Aggregates Appear Adjacent to Lysosomes.
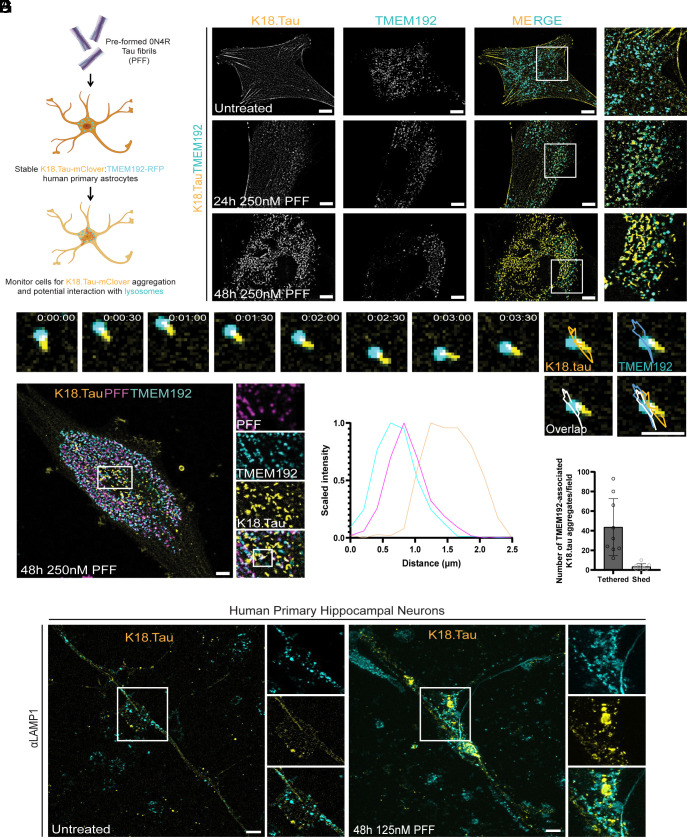
Although tau fibril endocytosis has been well documented, it is still unclear whether lysosome-associated fibrils are the species that trigger cytosolic tau aggregation (14, 15) (Fig. 4A). To address this question, we applied the astrocyte model of PFF endocytosis to study the effect of fibril uptake on the state of cytosolic tau (Fig. 4B). First, we transduced our TMEM192–RFP-expressing astrocytes with a mClover2-tagged truncated version of aggregation-sensitive tau (K18.tau) (12, 14). In the absence of fibrils, K18.tau was diffuse and/or localized to stress fibers and did not specifically interact with lysosomes or microtubules (Fig. 4B).
Fig. 4.
Soluble tau aggregation occurs in the proximity of lysosomes. To model the tau fibril uptake mechanism by astrocytes in vitro, we treated astrocytes stably expressing K18.tau-mClover and TMEM192–RFP with PFF (A). These dual astrocytes were used in live imaging experiments to determine what impact PFF endocytosis had on the cytoplasmic pool of tau (B). At rest, cells had diffuse K18.tau expression that did not interact with lysosomes (Top panel). After 24 h of fibril treatment, K18.tau aggregates could be detected in close proximity to TMEM192-positive lysosomes (Middle panel). The amount of K18 aggregation increased at 48 h posttreatment with numerous lysosomes found associated with large K18 aggregates (Lower panel). Representative trace of a lysosome-associated K18 aggregate during brief time-lapse imaging (C) (n = 393). Representative cell from the PFF seeding assay repeated with fluorescently labeled PFF to verify lysosomal accumulation and association with K18 aggregates (D) (n = 100). Fluorescence intensity plot of the line scan shown in D, revealing the spatial distribution of PFF, lysosomes, and K18 aggregates (E). (F) Quantification of the number of lysosome-tethered versus shed tau aggregates (n = 3 replicates, three random and independent fields of 20 cells per replicate, n = 60 cells). (G) K18.tau expression is diffuse in untreated primary human neurons but forms endogenous LAMP1-associated aggregates after PFF treatment (n = 3 replicates, 52 cells). [Scale bars: 10 µm (B, D, and G); 2 µm (C).]
The dual reporter astrocytes were then treated with 250 nM of PFF and monitored for aggregation for 1 to 2 d. K18.tau aggregates appeared adjacent to lysosomes within 24 h of incubation with fibrils (Fig. 4B). At 48 h post fibril uptake, K18.tau aggregates appeared to be elongated, resembling the tau tangles from those of tufted astrocytes (47) and interacted dynamically with lysosomes (Fig. 4C). Approximately 93% of lysosome-associated elongated K18.tau aggregates remained tethered to the lysosome surface over a 3-min window of observation (Fig. 4C). Imaging labeled PFF with K18.tau showed that the K18.tau aggregates extended from the exterior of lysosomes containing PFF (Fig. 4 D–F). We then investigated whether or not K18.tau aggregates could be found in proximity of lysosomes in neurons treated with PFF. To test this, neurons were transduced with K18.tau-mClover lentiviruses and treated 7 d later with 125 nM PFF for 48 h (Fig. 4G). In agreement with our findings in astrocytes, we observed K18.tau as a diffuse cytosolic signal in untreated neurons, while aggregates appeared after PFF treatment in the presence of neuron lysosomes labeled with endogenous LAMP1 antibody. These data strongly suggest that lysosomal PFF are responsible for nucleating the observed cytosolic tau aggregates.
Spatial Organization of Tau Aggregates on the Surface of Fibril-burdened Lysosomes.
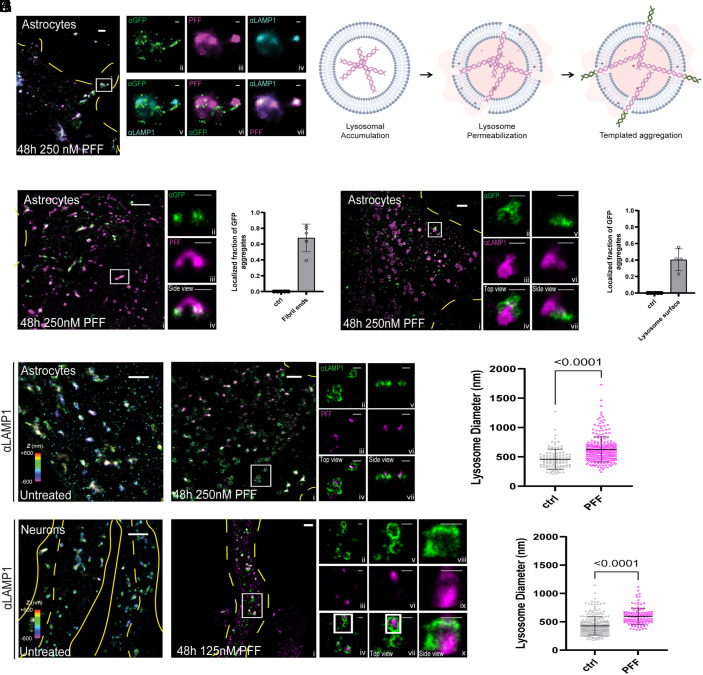
In order to resolve more precisely the spatial relationship between lysosomal PFF and cytosolic K18.tau aggregation, we employed multicolor STORM superresolution microscopy (48). Primary human astrocytes were transduced with the K18.tau-mClover fibril-induced aggregation sensor and treated with 647-labeled PFF for 48 h. Cells were stained for endogenous LAMP1, which was detected using a fluorescent secondary antibody with a CF680 label, and K18.tau-mClover was stained using anti-GFP antibodies with a CF583R secondary antibody. In the absence of PFF, there was no detectable GFP aggregation on the surface of LAMP-1-positive lysosomes. However, upon introduction of PFF into the media, fibril accumulation within LAMP-1-positive vesicles was readily apparent, and GFP aggregates were detected on the surface of the LAMP-1 fluorophore boundary (Fig. 5A and Movie S1). Our data suggest that lysosomal PFFs are responsible for the induced aggregation of K18.tau and that this aggregation occurs on the surface of lysosomes harboring PFF (Fig. 5B and Movie S2).
Fig. 5.
STORM imaging highlights interactions between lysosomes, added tau fibrils, and intracellularly expressed K18.tau-mClover. (A) 3-color STORM image of tau fibrils (magenta), lamp1 (cyan), and tau (green) in a human astrocyte that has been treated with PFF. The full image of the cell is shown on the left, and zoom-ins of the white-boxed region for each channel are shown on the Right. The Top panels are single-color images of each channel, and the Lower panels are two-color merges between channels. (B) Proposed model of PFF-induced templated aggregation on lysosome membranes. (C) Two-color, 3D-STORM images of 0N4R-treated cells staining intracellularly expressed K18.tau-mClover (green) and 0N4R fibrils (magenta). Side panels show a zoom-in of the white box region from a side view. (D) Histogram of puncta found at the tips of 0N4R fibrils. wo-color, 3D-STORM images of PFF-treated cells staining PFF (magenta) and lamp1 (green). Side panels show a zoom-in of the white box region. (E) Two-color, 3D-STORM images of PFF-treated cells staining for intracellularly expressed K18.tau-mClover (green) and lamp1 (magenta). Side panels show zoom-in of the white-box region from top (Left) and side (Right) views. (F) Histogram showing the number of K18.tau-mClover puncta found localized near the surface of the lamp1 lysosome signal. (G) 3D-STORM images of lamp1 in untreated astrocytes (Left) and astrocytes incubated with PFF (Right). Color encodes axial position (color bar, Left) and two color rendering (Right). (H) Histogram of lysosome diameters obtained from the data in G, compared by two-tailed unpaired t test. (I) 3D-STORM images of lamp1 in untreated human neurons (Left) and neurons incubated with PFF (Right). Color encodes axial position (color bar, Left) and two color rendering (Right). (J) Histogram of lysosome diameters obtained from the data in I, compared by two-tailed unpaired t test. Scale bars for large images (A–C, E, and H), 2 µm. Scale bars for smaller panels (A, B, E, F, H, and I), 500 nm.
To support our model, we next used 3D-STORM to characterize the interactions between K18.tau aggregates with fibrils and lysosomes (49). In agreement with our initial 3-color STORM observations, GFP aggregates could be detected on the ends of labeled PFF (Fig. 5 C and D). Additionally, approximately 68% and 40% of the GFP aggregates were localized to fibril ends and lysosome surfaces, respectively (Fig. 5 E and F). GFP aggregate density was detected primarily on the outside of the LAMP-1 sphere boundary, forming aggregates and extensions projecting away from the center of the lysosome body (Fig. 5 E and F). The addition of fibrils to the media also increased the diameter of astrocyte lysosomes by roughly 1.4-fold, from 460 nm to 626 nm (Fig. 5 G and H). We observed the same phenotype in neurons where lysosomes swelled from 428 nm to 596 nm after PFF treatment.
Discussion
In the prion-like propagation model for the spread of amyloid in the brain, key steps have been established, including the ability of tau seeds to be endocytosed and to nucleate aggregation of cytosolic tau (4). Yet, the mechanism for egress from the endosomal pathway to trigger cytosolic tau aggregation has remained mysterious. A nanoscale membrane damage mechanism for tau escape was suggested on the basis of a seeded tau aggregation phenotype of the ESCRT-III subunit CHMP6 (14), but the mechanism of action was not resolved. Here, we showed that lysosomes are deacidified following tau uptake, consistent with nanoscale membrane damage. While this could in principle have been explained by V-ATPase inhibition, we further found that the ESCRT-III subunits CHMP2B and CHMP4B were recruited to lysosomes under these conditions, diagnostic for nanoscale membrane damage. Moreover, we found cytosolic tau aggregates at the lysosome membrane surface and confirmed this event at high spatial resolution using STORM. Collectively, these data are hard to explain by alternative mechanisms. These data establish, at least under the tested conditions using recombinant PFFs in neurons and astrocytes, that tau escape occurs via nanoscale lysosomal membrane damage.
The finding of the role of ESCRTs in lysosomal membrane repair (22, 23) was followed more recently by characterization of the role of cholesterol and PI(4)P transfer from the ER (50, 51). Progress in the field has been driven primarily by the use of the model damaging agent LLOME. Recently, defects in the ESCRT pathway in its role as a lysosomal repair machine have been linked to Alzheimer’s disease (52). Now we found that tau fibrils are robust inducers of nanoscale lysosomal membrane damage and ESCRT recruitment, which will be important to characterize further as biomedically and physiologically relevant damaging agents.
The nanoscale permeabilization mechanism seems to be different from the well-established mechanism for viral pathogen escape from the endolysosomal system, fusion with the endolysosomal membrane (53). In contrast to the creation and resolution of a fusion pore by influenza or other viruses, this mechanism implies fibrillar seeds could protrude through the membrane into the cytosol, where they could template nucleation at the membrane surface, which is the predominant mechanism that we see. They could also break off, accounting for a subset of nucleation events that do not appear to be lysosome-adjacent. The observation of lysosomal membrane permeabilization begs the question of how tau interacts with membranes tightly enough to penetrate them. Unlike synuclein (54–56), there have not been extensive reports on how tau interacts with membranes in purified systems (57). One possiblity is that, as reported for Huntingtin (58), the bulky and rigid fibrils exert mechanical pressure on membranes without necessarily binding to them strongly.
Here, we found that lysosomal permeabilization and nanoscale damage, and lysosomal templating by tau, occurs in cultured astrocytes and neurons. The data do not establish how common this mechanism is in other systems and relative to other potential mechanisms, such as exosome (59) and tunneling nanotube (TNT)-dependent transfer (60, 61). Another limitation of this study is the use of recombinant PFFs whose structure differs from those found in patients with tauopathies (11). It will be exciting to explore whether this mechanism is more general and might apply to the spread of other forms of amyloid in the brain. These data raise questions about the fate of fibrillar cargo taken up by autophagy and lysosomes as a clearance response. If endocytosed fibrils can disrupt lysosomes, presumably fibrils taken up by autophagy could do the same. This highlights the importance of understanding the ESCRT-dependent damage response (Fig. 2), and potentially targeting it for upregulation as a therapeutic strategy to prevent spread. This could be of value both from the point of view of restricting cell-to-cell spread and making autophagic-lysosomal clearance as robust as possible.
Methods
Cell Culture and Cell Line Generation.
Human primary astrocytes were obtained from Sciencell (catalog #1800), and cryopreserved at the UCB Cell Culture Facility by A. Killilea. Human primary astrocytes used for experiments were maintained using Sciencell Astrocyte media (catalog #1801) for no more than 10 passages. Human primary hippocampal neurons were additionally acquired from Sciencell (catalog #1540) and maintained using Sciencell Neuron media (catalog #1521). All primary cells were grown on plates or glass coverslips coated with 2 µg/mL poly-L-Lysine (Sciencell catalog: 0403). HEK293T cells were cultured for no more than 20 passages in DMEM supplemented with 10% fetal bovine serum, Pen/Strep (Life Technologies, catalog number 15140122), and l-glutamine (Life Technologies, catalog number 25030081). All cells were maintained in a copper-lined Heracell VIOS 160i tissue culture incubator (ThermoFischer catalog: 51033574) at 37 °C and 5% CO2 and checked for Mycoplasma contamination.
To generate K18.tau-mClover, TMEM192–RFP, pHLARE, CHMP2B–HALO, CHMP2Bintron5–HALO, CHMP4B–HALO, Gal3–GFP, dCas9–KRAB, sgScramble, and sgCHMP6 lentiviruses, 2E6 HEK293T cells were seeded into 10 cm plates and transfected the next day with 45 µL Mirus LT1 transfection reagent (MIR2300) added to a mixture of 5 µg each (15 µg total) of plasmids VSV-G (addgene: 8454), R8.74 (addgene: 22036), and one of the following: K18.tau-mClover (addgene: 133058), TMEM192–RFP (addgene: 134631), pHLARE (addgene: 164478), CHMP2B–HALO, CHMP2B–HALO, CHMP2Bintron5–HALO (this study), Gal3–GFP (addgene: 62734), sgScramble-BFP (addgene: 128749), dCas9-KRAB, or sgCHMP6 (Martin Kampmann lab) in 1.5 mL Optimem (ThermoFischer catalog: 31985062) according to the manufacturer’s recommendations. The supernatant containing viruses was obtained 3 d posttransfection, clarified by centrifugation at 2,500 rpm for 2.5 min and concentrated 10-fold using Lenti-X concentrator (Takara Bio catalog: 631231). On the day prior to transduction, astrocytes were seeded at a density of 100,000 cells per well into individual wells of a 12-chamber plate (catalog number: 07-200-82). Human primary hippocampal neurons were transduced in the same manner. Stable pools of astrocytes expressing desired proteins of interest were obtained by titrating virus concentrate to achieve near 100% expression efficiency, and passaging cells once prior to experiments.
Production and Fibrilization of Recombinant 0N4R PFF for Cell Seeding Assays.
Generation of recombinant 0N4R PFF was based on a previous report (14). Briefly, N-terminal 6× His tagged Human WT 0N4R tau protein is expressed in RosettaTM 2 (DE3)- competent cells in LB medium supplemented with Ampicillin (100 μg/mL), induced at 0.6 OD with 1 mM isopropyl 1-thio-β-d-galactopyranoside at 37 °C for 3 h. After lysis through tip sonication in Buffer A (1× PBS, pH 7.5, 2 mM MgCl2, 10 mM EGTA, 1 mM TCEP), supplemented with 0.1 mM PMSF and two EDTA free protease inhibitor tablets (Roche, Basel, Switzerland), the expressed protein was extracted from the supernatant using PierceTM High-Capacity Ni-IMAC Resin (Thermo Scientific) and eluted using lysis buffer supplemented with 300 mM Imidazole pH 7.5. TEV protease was added to the IMAC elute and subjected to overnight dialysis in Buffer B (20 mM MES, pH 6.8, 50 mM NaCl, 1 mM EGTA, 1 mM MgCl2, 1 mM TCEP). The dialyzed protein was purified by cation exchange chromatography using 5 mL HiTrap SP (Cytiva) and eluted with a linear gradient of 0 to 1 M NaCl. Fractions containing tau as determined by Coomassie-stained SDS-PAGE were loaded onto the Superdex 75 16/60 column (Cytiva) for gel filtration and eluted using SEC buffer (1× PBS, pH 7.5, 2 mM MgCl2, 1 mM TCEP). The concentration of the purified protein was calculated by measuring the absorbance at 280 nm. Finally, the protein was concentrated at approximately 15 μM, snap-frozen, and stored at −80 °C in 75 μL aliquots.
Aggregation of monomeric tau into PFF was induced by incubating 10 μM tau with 1.3 μM of heparin (Average molecular weight ~ 16,500 g/mol) (Thermo Scientific) and shaken at 37 °C overnight at 1,200 rpm. The resulting tau fibrils were fluorescently labeled with AlexaFluor 647 NHS-ester dye (Invitrogen) following the manufacturer’s protocol. Briefly, NaHCO3 was added to a final concentration of 100 μM to raise the pH of labeling mixture before adding the dye to molar (monomeric) protein to dye ratio of 3:1. This fibril–dye mixture was kept in dark at room temperature for 1 h before removing the unlabeled dye with a Zeba 7k MWCO spin desalting column (Thermo Scientific).
For quality control, PFF were negatively stained with 0.8% (w/v) uranyl formate (pH 5.5 to 6.0) on thin amorphous carbon-layered 400-mesh copper grids (Electron Microscopy Services). Five μL of sample was applied to the grid for 1 min followed by one wash with 6 μL of uranyl formate and a 1 min incubation with 6 μL of uranyl formate, removed with WhatmanTM filter paper (Cytiva). Grids were imaged at room temperature using a Fei Tecnai 12 microscope operating at 120 kV. Images were acquired on a 4,000 TVIPS TemCam-F416 camera at ×30,000 resulting in a sampling of 3.70 Å/pixel. PFF were typically greater than 10 µm in length but could be broken up into 50 nm fragments using a continual sonication water bath at 37 degrees for 30 min, verified by negative stain as above. Sonicated PFF were used in cell seeding experiments by adding the fibrils directly to the media at a final concentration of 250 nM (unless otherwise indicated) and monitoring cells for 24 to 48 h.
Monitoring Lysosomal pH.
pHLARE expressing astrocytes were seeded at a density of 100,000 cells per well onto poly-L-Lysine (Sigma catalog MILL-A-005-C) coated coverslips in individual wells of a 12-chamber plate. Bafilomycin (Sigma catalog: 19-600-010UG) and PFF treatment was conducted at the indicated concentrations and durations prior to fixing cells with 4% Paraformaldehyde (EMS catalog 15710-S) in DPBS (Corning catalog: 21040CV) for 15 min. Fixed coverslips were then mounted onto glass slides using ProLong Gold with DAPI (ThermoFischer catalog: P36935) and left to harden in the dark overnight prior to imaging. Cells were imaged using a using a Nikon A1 confocal microscope with a 63× Plan Apochromat 1.4 numerical aperture objective with identical imaging settings across replicates. Diffuse GFP signal was subtraced and cell images were processed using a median filter of seven pixels in ImageJ prior to quantification. Total RFP area per cell was then quantified and compared across cells before quantifying the overlapping GFP-positive area using the ImageJ Coloc2 Plugin (https://imagej.net/plugins/coloc-2). Two-way ANOVA was performed to compare the effect of Bafilomycin and PFF on lysosomal pH. Coloc2 was additionally used to compare GFP-positive lysosomes to GFP-positive PFF in cells treated with labeled fibrils. Linear regression was performed on the Mander’s coefficient obtained from Coloc2 analysis of GFP-positive lysosomes (x axis) and GFP-positive PFF (y axis).
Visualization of ESCRT and Gal3 Recruitment to Lysosomes.
TMEM192–RFP expressing astrocytes were fisrt treated with fluorescently labeled PFF to verify lysosomal accumulation. These cells were additionally transduced with CHMP2B–HALO, CHMP2Bintron5–HALO, or CHMP4B–HALO in parallel experiments for the monitoring of ESCRT responses to PFF treatment. CHMP4B signal was visualized using a secondary promoter driving BFP expression or far-red HALO ligand according to the manufacturer’s recommendations (Promega catalog GA1120). Cells were processed as above for imaging. Number of cells expressing CHMP4B puncta was determined by scanning fields of cells positive for CHMP4B signal and manaully reporting the fraction of cells exhibiting puncta.
TMEM192–RFP expressing astrocytes were also transduced with Gal3–GFP and subjected to the same fibril treatment as CHMP4B-expressing cells. Gal3–GFP recruitment to lysosomes was monitored by TMEM192–RFP colocalization. Gal3-positive TMEM192–RFP area was measured before and after treatment using Coloc2 and normalized to resting levels of Gal3-positive lysosomes. Treated cells were compared to resting cells using a two-tailed unpaired t test. Cell images were processed as above for quantification and visualization.
Interference with the ESCRT Machinery and Monitoring of Lysosome Permeabiltiy.
Astrocytes expressing Gal3–GFP and TMEM192–RFP were seeded onto poly-L-Lysine coated glass coverslips in 12-well plates prior to mock transduction or transduction with either CHMP2B–HALO or CHMP2Bintron5–HALO lentiviruses. Two days posttransduction, cells were treated with PFF for an additional 2 d before being fixed and assessed for increases in Gal3–GFP recruitment to TMEM192–RFP containing lysosomes. ESCRT-expressing cells were identified by BFP expression and used for analysis. A Pearson correlation coefficient of Gal3–GFP and TMEM192–RFP signals was determined for each sample and compared by one-way ANOVA.
To perform CRISPRi-mediated knockdown of CHMP6, a stable astrocyte cell line expressing Gal3–GFP and TMEM192–RFP was additionally transduced with dCas9-KRAB-BFP until achieving nearly 100% transduction efficiency as measured by BFP puncta signal. CRISPRi-competent cells were then expanded and passaged prior to experiments. Cells were prepared for imaging experiments as above and transduced with either sgScramble-BFP lentiviruses or sgCHMP6 lentiviruses with a small amount of sgScramble-BFP lentiviruses to identify cells expressing sgRNAs via diffuse cytoplasmic BFP signal, for 3 d. On day 3, cells were treated with PFF for two additional days before sample preparation and processing. Cytoplasmic BFP signal was used to guide image acquisiton and analysis. Pearson correlation coefficients were determined as above and compared by a two-tailed unpaired t test.
Live Cell Imaging and Lysosome Tracking.
Astrocytes expressing K18.tau-mClover and TMEM192–RFP were seeded into poly-L-Lysine coated 35-mm glass bottom dishes (no. P35G-1.5-14-C, MatTek). Cells were treated with unlabeled PFF for 24 and 48 h prior to conducting live imaging experiments. Timelapse imaging was conducted every 15 s for 3 min to track lysosomes with adjacent K18.tau aggregates. The centroid of the overlapping fluorescenct region was monitored over time using the ImageJ Particle Tracker Plugin (https://imagej.net/plugins/particle-tracker). The centroid trace was then overlayed on the last frame of the timelapse. Cell images were processed as above for quantification and visualization.
Superresolution Imaging of PFF-treated Astrocytes.
For STORM imaging, astrocytes loaded with either unlabeled or AF647 NHS ester-labeled tau fibrils (0N4R PFF, described earlier in methods) were fixed with 3% (vol/vol) paraformaldehyde (Electron Microscopy Sciences, 15714) and 0.1% (vol/vol) glutaraldehyde (Electron Microscopy Sciences, 16365) in DPBS for 20 min and then washed twice with a freshly prepared 0.1% NaBH4 solution followed by three additional washes with DPBS. Cells were then incubated with a solution containing the membrane pore-forming protein Streptolysin O (SLO) (Sigma catalog: S5265-25KU) sufficient to permeabilize 90% of HeLa cells, as used previously (62). SLO-permeabilized cells were washed twice with DPBS followed by incubation with the antibodies of interest, with SLO pores allowing antibodies to pass through cellular membranes while keeping their structure intact. For the two-color images, LAMP1 was initially targeted with mouse anti-LAMP1 (Santa Cruz Biotech, SC-20011) followed by either donkey anti-mouse CF583R (Biotium catalog: 20794-500UL) for samples loaded with labeled fibrils or goat anti-mouse AF647 (Thermo-Fischer, A21235) for samples loaded with unlabeled fibrils. This same approach was used to label lysosomes in neurons expressing K18.tau-mClover. K18.tau-mClover was targeted with mouse anti-GFP (Thermo-Fischer, MA5-15256), followed by donkey anti-mouse CF583R (Biotium catalog: 20794-500UL). Three-color STORM used the following labeling scheme: AF647 NHS ester labeled PFF, while donkey anti-rabbit CF680 (Biotium catalog: 20418-50UL) and donkey anti-mouse CF583R (Biotium catalog: 20794-500UL), were used as secondaries for LAMP1 (rabbit anti-LAMP1, Invitrogen, PA1654A) and GFP (mouse anti-GFP, Thermo-Fischer, MA5-15256), respectively. SLO-permeabilized cells were blocked for 30 min with DPBS containing 3% BSA prior to antibody incubation. All primary antibodies were prepared at a dilution of 1:100 and secondary antibodies at a 1:500 dilution, in DPBS containing 3% BSA. Cells were incubated with primary antibodies for 1 h at room temp, washed three times with DPBS between primary and secondary incubations, and incubated with secondary antibodies for 1 h at room temp in the dark. Residual secondary antibody was removed by three additional washes with DPBS.
Two-color 3D-STORM was performed for samples labeled by Alexa Fluor 647 and CF583R on a homebuilt setup based on a Nikon Eclipse Ti-U inverted microscope, as previously described (63, 64). The dye-labeled samples were mounted with a Tris⋅HCl–based imaging buffer containing 5% (wt/vol) glucose, 100 mM cysteamine (Sigma-Aldrich, 30070), 0.8 mg/mL glucose oxidase (Sigma-Aldrich, G2133), and 40 µg/mL catalase (Sigma-Aldrich, C30). The sample was sequentially STORM-imaged for Alexa Fluor 647 and CF583R with 647- and 560-nm illuminations at ∼2 kW/cm2. The angle of incidence was slightly smaller than the critical angle of total internal reflection, thus illuminating a few micrometers into the sample. A weak (0 to 1 W/cm2) 405-nm laser was applied to assist photoswitching. A cylindrical lens was inserted into the optical path for 3D localization (49). The resulting stochastic photoswitching of single-molecule fluorescence was recorded using an Andor iXon Ultra 897 EM-CCD camera at 110 frames per second, for ∼80,000 frames per STORM image. The raw STORM data were analyzed using previously described methods (49).
Three-color STORM was performed on another homebuilt setup based on a Nikon Eclipse Ti-U inverted microscope, as previously described (65). Samples with Alexa Fluor 647 and CF680 were first simultaneously excited under 647-nm illumination and distinguished through ratiometric single-molecule detection in two spectrally split wide-field channels (66) using a dichroic mirror (Chroma T685lpxr), and CF583R was subsequently imaged under 560-nm excitation.
To generate movies of 3D STORM data, The STORM datasets were loaded and processed as described above. Isolated examples of astrocyte PFF and lysosomes were displayed in three dimensions. Images of the regions were saved in 2-degree increments for a full rotation and then combined in ImageJ to create the final 3D rotation movies.
Statistical Measurements.
Cell data were plotted and analyzed using Graphpad Prism 9.0. Linear regression, two-tailed unpaired t tests, or one- and two-way ANOVA were used to compare experimental samples when appropriate and as indicated. Statistical significance is indicated by plotting P values to four digits.
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
A 3D STORM example of a lysosome (magenta) with K18.tau aggregates (green) from PFF-treated astrocytes is shown and rotated.
A 3D STORM example of a PFF (magenta) with K18.tau aggregates (green) from PFF-treated astrocytes is shown and rotated.
Acknowledgments
We thank Senthil Kaniyappan for comments on the manuscript. This research was supported by Hoffmann-La Roche as part of the Alliance for Therapies in Neuroscience (J.H.H.), NIH/NIA R01 AG062359 (M.K.), NIH/NIGMS R35GM149349 (K.X.), and the NSF Graduate Research Fellowship DGE 2146752 (T. J.). J.H.H. receives research funding from Genentech and Hoffmann-La Roche.
Author contributions
K.R., T.J., A.M.-R., M.K., K.X., and J.H.H. designed research; K.R., T.J., and A.M.-R. performed research; K.R. and S.S. contributed new reagents/analytic tools; K.R., T.J., and A.M.-R. analyzed data; and K.R. and J.H.H. wrote the paper.
Competing interests
J.H.H. is a co-founder and shareholder of Casma Therapeutics. M.K. serves on the Scientific Advisory Boards of Engine Biosciences, Casma Therapeutics, Cajal Neuroscience, Alector, and Montara Therapeutics, and is an advisor to Modulo Bio and Recursion Therapeutics. J.H.H. is a shareholder of Casma Therapeutics.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
All study data are included in the article and/or supporting information.
Supporting Information
References
- 1.Riek R., Eisenberg D. S., The activities of amyloids from a structural perspective. Nature 539, 227–235 (2016), 10.1038/nature20416. [DOI] [PubMed] [Google Scholar]
- 2.Peng C., Trojanowski J. Q., Lee V. M., Protein transmission in neurodegenerative diseases. Nat. Rev. Neurol. 16, 199–212 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goedert M., Clavaguera F., Tolnay M., The propagation of prion-like protein inclusions in neurodegenerative diseases. Trends Neurosci. 33, 317–325 (2010), 10.1016/j.tins.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 4.Vaquer-Alicea J., Diamond M. I., Propagation of protein aggregation in neurodegenerative diseases. Annu. Rev. Biochem. 88, 785–810 (2019), 10.1146/annurev-biochem-061516-045049. [DOI] [PubMed] [Google Scholar]
- 5.Wang Y. P., Mandelkow E., Tau in physiology and pathology. Nat. Rev. Neurosci. 17, 5–21 (2016), 10.1038/nrn.2015.1. [DOI] [PubMed] [Google Scholar]
- 6.Müller R., Heinrich M., Heck S., Blohm D., Richter-Landsberg C., Expression of microtubule-associated proteins MAP2 and tau in cultured rat brain oligodendrocytes. Cell Tissue Res. 288, 239–249 (1997), 10.1007/s004410050809. [DOI] [PubMed] [Google Scholar]
- 7.de Calignon A., et al. , Propagation of tau pathology in a model of early Alzheimer’s disease. Neuron 73, 685–697 (2012), 10.1016/j.neuron.2011.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Piras A., Collin L., Grüninger F., Graff C., Rönnbäck A., Autophagic and lysosomal defects in human tauopathies: Analysis of post-mortem brain from patients with familial Alzheimer disease, corticobasal degeneration and progressive supranuclear palsy. Acta Neuropathol. Commun. 4, 22 (2016), 10.1186/s40478-016-0292-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richetin K., et al. , Tau accumulation in astrocytes of the dentate gyrus induces neuronal dysfunction and memory deficits in Alzheimer’s disease. Nat. Neurosci. 23, 1567–1579 (2020), 10.1038/s41593-020-00728-x. [DOI] [PubMed] [Google Scholar]
- 10.Scheres S. H., Zhang W., Falcon B., Goedert M., Cryo-EM structures of tau filaments. Curr. Opin. Struct. Biol. 64, 17–25 (2020), 10.1016/j.sbi.2020.05.011. [DOI] [PubMed] [Google Scholar]
- 11.Shi Y., et al. , Structure-based classification of tauopathies. Nature 598, 359–363 (2021), 10.1038/s41586-021-03911-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frost B., Jacks R. L., Diamond M. I., Propagation of tau misfolding from the outside to the inside of a cell. J. Biol. Chem. 284, 12845–12852 (2009), 10.1074/jbc.M808759200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saha I., et al. , The AAA+ chaperone VCP disaggregates Tau fibrils and generates aggregate seeds in a cellular system. Nat. Commun. 14, 560 (2023), 10.1038/s41467-023-36058-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen J. J., et al. , Compromised function of the ESCRT pathway promotes endolysosomal escape of tau seeds and propagation of tau aggregation. J. Biol. Chem. 294, 18952–18966 (2019), 10.1074/jbc.RA119.009432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kolay S., et al. , The dual fates of exogenous tau seeds: Lysosomal clearance versus cytoplasmic amplification. J. Biol. Chem. 298, 102014 (2022), 10.1016/j.jbc.2022.102014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thurston T. L., Wandel M. P., von Muhlinen N., Foeglein A., Randow F., Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature 482, 414–418 (2012), 10.1038/nature10744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Papadopoulos C., et al. , VCP/p97 cooperates with YOD1, UBXD1 and PLAA to drive clearance of ruptured lysosomes by autophagy. Embo J. 36, 135–150 (2017), 10.15252/embj.201695148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Polanco J. C., Hand G. R., Briner A., Li C., Götz J., Exosomes induce endolysosomal permeabilization as a gateway by which exosomal tau seeds escape into the cytosol. Acta Neuropathol. 141, 235–256 (2021), 10.1007/s00401-020-02254-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Calafate S., Flavin W., Verstreken P., Moechars D., Loss of Bin1 promotes the propagation of tau pathology. Cell Rep. 17, 931–940 (2016), 10.1016/j.celrep.2016.09.063. [DOI] [PubMed] [Google Scholar]
- 20.Hurley J. H., ESCRTs are everywhere. EMBO J. 34, 2398–2407 (2015), 10.15252/embj.201592484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vietri M., Radulovic M., Stenmark H., The many functions of ESCRTs. Nat. Rev. Mol. Cell Biol. 21, 25–42 (2020), 10.1038/s41580-019-0177-4. [DOI] [PubMed] [Google Scholar]
- 22.Radulovic M., et al. , ESCRT-mediated lysosome repair precedes lysophagy and promotes cell survival. EMBO J. 37, e99753 (2018), 10.15252/embj.201899753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Skowyra M. L., Schlesinger P. H., Naismith T. V., Hanson P. I., Triggered recruitment of ESCRT machinery promotes endolysosomal repair. Science 360, eaar5078 (2018), 10.1126/science.aar5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bohannon K. P., Hanson P. I., ESCRT puts its thumb on the nanoscale: Fixing tiny holes in endolysosomes. Curr. Opin. Cell Biol. 65, 122–130 (2020), 10.1016/j.ceb.2020.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhen Y., Radulovic M., Vietri M., Stenmark H., Sealing holes in cellular membranes. EMBO J. 40, e106922 (2021), 10.15252/embj.2020106922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shukla S., Larsen K. P., Ou C., Rose K., Hurley J. H., In vitro reconstitution of calcium-dependent recruitment of the human ESCRT machinery in lysosomal membrane repair. Proc. Natl. Acad. Sci. U.S.A. 119, e2205590119 (2022), 10.1073/pnas.2205590119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reid M. J., Beltran-Lobo P., Johnson L., Perez-Nievas B. G., Noble W., Astrocytes in tauopathies. Front. Neurol. 11, 572850 (2020), 10.3389/fneur.2020.572850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gomez-Arboledas A., et al. , Phagocytic clearance of presynaptic dystrophies by reactive astrocytes in Alzheimer’s disease. Glia 66, 637–653 (2018), 10.1002/glia.23270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perea J. R., et al. , Extracellular monomeric tau is internalized by astrocytes. Front. Neurosci. 13, 442 (2019), 10.3389/fnins.2019.00442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fleeman R. M., Proctor E. A., Astrocytic propagation of tau in the context of Alzheimer’s disease. Front. Cell Neurosci. 15, 645233 (2021), 10.3389/fncel.2021.645233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chung W. S., Allen N. J., Eroglu C., Astrocytes control synapse formation, function, and elimination. Cold Spring Harb. Perspect. Biol. 7, a020370 (2015), 10.1101/cshperspect.a020370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Webb B. A., et al. , pHLARE: A new biosensor reveals decreased lysosome pH in cancer cells. Mol. Biol. Cell 32, 131–142 (2021), 10.1091/mbc.E20-06-0383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ye C. Y., et al. , Tau overload associated insufficient lysosomal hydrolysis activity through deacidification of lysosomes. Research Square [Preprint] (2023). 10.21203/rs.3.rs-3294833/v1 (Accessed 4 September 2023). [DOI]
- 34.Kim S.-H., et al. , Endolysosomal impairment by binding of amyloid beta or MAPT/Tau to V-ATPase and rescue via the HYAL-CD44 axis in Alzheimer disease. Autophagy 19, 2318–2337 (2023), 10.1080/15548627.2023.2181614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Drummond E., et al. , Phosphorylated tau interactome in the human Alzheimer’s disease brain. Brain 143, 2803–2817 (2020), 10.1093/brain/awaa223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu J. W., et al. , Small misfolded Tau species are internalized via bulk endocytosis and anterogradely and retrogradely transported in neurons. J. Biol. Chem. 288, 1856–1870 (2013), 10.1074/jbc.M112.394528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Evans L. D., et al. , Extracellular monomeric and aggregated tau efficiently enter human neurons through overlapping but distinct pathways. Cell Rep. 22, 3612–3624 (2018), 10.1016/j.celrep.2018.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gibbons G. S., et al. , GFP-Mutant human tau transgenic mice develop tauopathy following CNS injections of Alzheimer’s brain-derived pathological tau or synthetic mutant human tau fibrils. J. Neurosci. 37, 11485–11494 (2017), 10.1523/jneurosci.2393-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mierzwa B. E., et al. , Dynamic subunit turnover in ESCRT-III assemblies is regulated by Vps4 to mediate membrane remodelling during cytokinesis. Nat. Cell Biol. 19, 787–798 (2017), 10.1038/ncb3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Skibinski G., et al. , Mutations in the endosomal ESCRTIII-complex subunit CHMP2B in frontotemporal dementia. Nat. Genet. 37, 806–808 (2005), 10.1038/ng1609. [DOI] [PubMed] [Google Scholar]
- 41.Parkinson N., et al. , ALS phenotypes with mutations in CHMP2B (charged multivesicular body protein 2B). Neurology 67, 1074–1077 (2006), 10.1212/01.wnl.0000231510.89311.8b. [DOI] [PubMed] [Google Scholar]
- 42.Yang B., Stjepanovic G., Shen Q., Martin A., Hurley J. H., Vps4 disassembles an ESCRT-III filament by global unfolding and processive translocation. Nat. Struct. Mol. Biol. 22, 492–498 (2015), 10.1038/nsmb.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.West R. J. H., Ugbode C., Gao F. B., Sweeney S. T., The pro-apoptotic JNK scaffold POSH/SH3RF1 mediates CHMP2BIntron5-associated toxicity in animal models of frontotemporal dementia. Hum. Mol. Genet. 27, 1382–1395 (2018), 10.1093/hmg/ddy048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van der Zee J., et al. , CHMP2B C-truncating mutations in frontotemporal lobar degeneration are associated with an aberrant endosomal phenotype in vitro. Hum. Mol. Genet. 17, 313–322 (2008), 10.1093/hmg/ddm309. [DOI] [PubMed] [Google Scholar]
- 45.Urwin H., et al. , Disruption of endocytic trafficking in frontotemporal dementia with CHMP2B mutations. Hum. Mol. Genet. 19, 2228–2238 (2010), 10.1093/hmg/ddq100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.West R. J. H., Ugbode C., Fort-Aznar L., Sweeney S. T., Neuroprotective activity of ursodeoxycholic acid in CHMP2B(Intron5) models of frontotemporal dementia. Neurobiol. Dis. 144, 105047 (2020), 10.1016/j.nbd.2020.105047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oshima K., Dickson D. W., Cortical Alzheimer type pathology does not influence tau pathology in progressive supranuclear palsy. Int. J. Clin. Exp. Pathol. 2, 399–406 (2009). [PMC free article] [PubMed] [Google Scholar]
- 48.Rust M. J., Bates M., Zhuang X., Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods 3, 793–796 (2006), 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang B., Wang W., Bates M., Zhuang X., Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science 319, 810–813 (2008), 10.1126/science.1153529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Radulovic M., et al. , Cholesterol transfer via endoplasmic reticulum contacts mediates lysosome damage repair. EMBO J. 41, e112677 (2022), 10.15252/embj.2022112677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tan J. X., Finkel T., A phosphoinositide signalling pathway mediates rapid lysosomal repair. Nature 609, 815–821 (2022), 10.1038/s41586-022-05164-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chou C. C., et al. , Proteostasis and lysosomal quality control deficits in Alzheimer’s disease neurons. bioRxiv [Preprint] (2023). 10.1101/2023.03.27.534444 (Accessed 10 April 2023). [DOI]
- 53.Harrison S. C., Viral membrane fusion. Nat. Struct. Mol. Biol. 15, 690–698 (2008), 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Braun A. R., et al. , α-Synuclein induces both positive mean curvature and negative gaussian curvature in membranes. J. Am. Chem. Soc. 134, 2613–2620 (2012), 10.1021/ja208316h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Makasewicz K., et al. , Cooperativity of α-synuclein binding to lipid membranes. ACS Chem. Neurosci. 12, 2099–2109 (2021), 10.1021/acschemneuro.1c00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Musteikytė G., et al. , Interactions of α-synuclein oligomers with lipid membranes. Biochim. Biophy. Acta 1863, 183536 (2021), 10.1016/j.bbamem.2020.183536. [DOI] [PubMed] [Google Scholar]
- 57.Majewski J., et al. , Lipid membrane templated misfolding and self-assembly of intrinsically disordered tau protein. Sci. Rep. 10, 13324 (2020), 10.1038/s41598-020-70208-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bäuerlein F. J. B., et al. , In situ architecture and cellular interactions of PolyQ inclusions. Cell 171, 179–187.e110 (2017), 10.1016/j.cell.2017.08.009. [DOI] [PubMed] [Google Scholar]
- 59.Polanco J. C., Scicluna B. J., Hill A. F., Gotz J., Extracellular vesicles isolated from the brains of rTg4510 mice seed tau protein aggregation in a threshold-dependent manner. J. Biol. Chem. 291, 12445–12466 (2016), 10.1074/jbc.M115.709485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Abounit S., Wu J. W., Duff K., Victoria G. S., Zurzolo C., Tunneling nanotubes: A possible highway in the spreading of tau and other prion-like proteins in neurodegenerative diseases. Prion 10, 344–351 (2016), 10.1080/19336896.2016.1223003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tardivel M., et al. , Tunneling nanotube (TNT)-mediated neuron-to neuron transfer of pathological Tau protein assemblies. Acta Neuropathol. Commun. 4, 117 (2016), 10.1186/s40478-016-0386-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Teng K. W., et al. , Labeling proteins inside living cells using external fluorophores for microscopy. Elife 5, e20378 (2016), 10.7554/eLife.20378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang M., Kenny S. J., Ge L., Xu K., Schekman R., Translocation of interleukin-1β into a vesicle intermediate in autophagy-mediated secretion. Elife 4, e11205 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang B., et al. , Transforming rhodamine dyes for (d)STORM super-resolution microscopy via 1,3-disubstituted imidazolium substitution. Angew. Chem. Int. Ed. Engl. 61, e202113612 (2022), 10.1002/anie.202113612. [DOI] [PubMed] [Google Scholar]
- 65.Ge L., et al. , Remodeling of ER-exit sites initiates a membrane supply pathway for autophagosome biogenesis. EMBO Rep. 18, 1586–1603 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bossi M., et al. , Multicolor far-field fluorescence nanoscopy through isolated detection of distinct molecular species. Nano Lett. 8, 2463–2468 (2008), 10.1021/nl801471d. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
A 3D STORM example of a lysosome (magenta) with K18.tau aggregates (green) from PFF-treated astrocytes is shown and rotated.
A 3D STORM example of a PFF (magenta) with K18.tau aggregates (green) from PFF-treated astrocytes is shown and rotated.
Data Availability Statement
All study data are included in the article and/or supporting information.