Abstract
Management of chronic obesity-associated metabolic disorders is a key challenge for biomedical researchers. During chronic obesity, visceral adipose tissue (VAT) undergoes substantial transformation characterized by a unique lipid-rich hypoxic AT microenvironment which plays a crucial role in VAT dysfunction, leading to insulin resistance (IR) and type 2 diabetes. Here, we demonstrate that obese AT microenvironment triggers the release of miR-210-3p microRNA-loaded extracellular vesicles from adipose tissue macrophages, which disseminate miR-210-3p to neighboring adipocytes, skeletal muscle cells, and hepatocytes through paracrine and endocrine actions, thereby influencing insulin sensitivity. Moreover, EVs collected from Dicer-silenced miR-210-3p–overexpressed bone marrow–derived macrophages induce glucose intolerance and IR in lean mice. Mechanistically, miR-210-3p interacts with the 3′-UTR of GLUT4 mRNA and silences its expression, compromising cellular glucose uptake and insulin sensitivity. Therapeutic inhibition of miR-210-3p in VAT notably rescues high-fat diet–fed mice from obesity-induced systemic glucose intolerance. Thus, targeting adipose tissue macrophage–specific miR-210-3p during obesity could be a promising strategy for managing IR and type 2 diabetes.
Keywords: obesity, adipose tissue microenvironment, extracellular vesicles, miR-210-3p, GLUT4, insulin resistance
Chronic obesity has emerged as a global epidemic of multifactorial poor health conditions with increasing risk of various metabolic disorders, including insulin resistance (IR) and type 2 diabetes (T2D) (1, 2). The obesity-associated systemic metabolic dysfunction influences cellular cross-talk within the adipose tissue microenvironment (ATenv) that critically regulates chronic low-grade inflammation in adipose tissue (AT) (3, 4). Despite extensive research efforts, effective therapeutic interventions targeting obesity-related metabolic dysregulation remain a pressing need. Obesity triggers substantial changes in AT, establishing a unique lipid-rich hypoxic ATenv (5, 6). This pathophysiological ATenv accelerates infiltration of immune cells, particularly monocytes, which differentiate into AT macrophages (ATMs) and switch their polarity from anti-inflammatory to proinflammatory state exacerbating AT inflammation and IR by activating the NF-κB pathway (7, 8). Recent single-cell transcriptomic dataset analyses have identified ATMs in obese individuals predominantly of Trem2+ lipid-associated macrophages, CD9+ macrophages, and other inflammatory macrophages (9, 10, 11), all of which play prominent roles in chronic AT inflammation and IR (12).
Extracellular vesicles (EVs) are small membrane-bound vesicles secreted by the cells including exosomes (30–120 nm) and microvesicles (150–1000 nm) that transfer a wide range of biomolecules to exert their biological functions (13, 14). Research in this direction demonstrated that EVs play a key role in exchanging organelles and various biomolecules, including miRNAs (15, 16, 17). A recent study revealed that exosomes containing miR-155, secreted by obese ATMs, contribute to IR by targeting peroxisome proliferator-activated receptor gamma (PPARγ), which regulates adipogenesis and indirectly influences the insulin signaling pathway (18). Additionally, AT-derived exosomal miR-27b has been implicated in vascular endothelial cell dysfunction, promoting atherogenesis by silencing PPARγ (19). To date, there is no evidence indicating that environmentally-induced miRNAs (hypoxamiRs) from obese ATMs have the capability to directly target molecules in the insulin signaling pathway leading to systemic insulin resistance.
The present study, for the first time, elucidates the direct involvement of obese ATenv-induced ATMs-derived miR-210-3p on the impairment of systemic insulin sensitivity and glucose homeostasis through silencing insulin signaling pathway molecule, GLUT4. ATMs dispense miR-210-3p to neighboring adipocytes through EVs in the lipid-rich hypoxic ATenv, which causes IR and glucose intolerance by targeting GLUT4 expression. We also found that miR-210-3p–enriched EVs, secreted from obese ATMs, notably inhibit insulin sensitivity in the distant skeletal muscle cells and hepatocytes via endocrine actions. Therefore, specific inhibition of miR-210-3p in ATMs and/or insulin target cells could serve as novel therapeutic strategies for managing obese-induced IR.
Results
Obesity induces enrichment of miR-210-3p in ATMs-derived EVs and its delivery to adipocytes
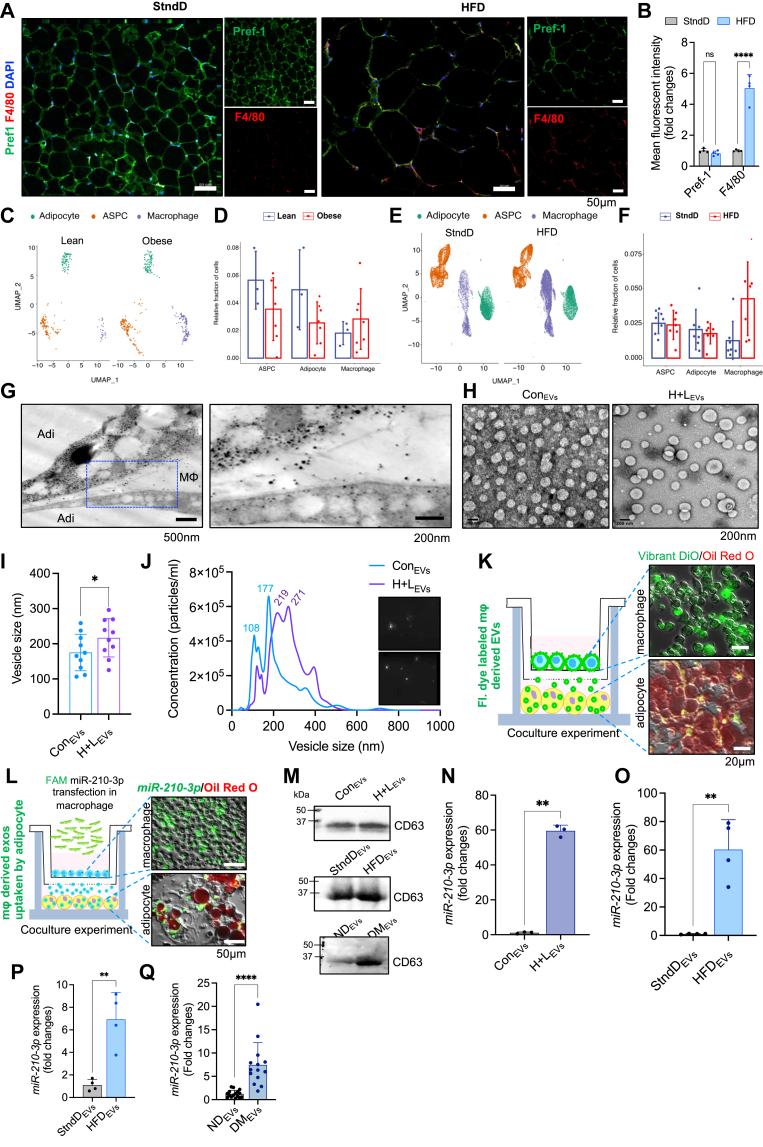
Along with others (12, 18, 20, 21), we also noted an increased quantum of macrophage population in the visceral adipose tissue (VAT) of high-fat diet (HFD)-fed mice compared to standard diet (StndD)-fed mice (Fig. 1, A and B), determined by staining tissue sections with F4/80 (macrophage marker) and Pref-1 (preadipocyte marker). Reanalysis of the publicly available single nuclei sequencing dataset of VAT from lean and obese human subjects as well as StndD and HFD mice (GSE176171) (22) revealed a significant increase in ATM populations both in the VAT samples of obese human (Fig. 1, C and D) and HFD mice (Fig. 1, E and F). We previously reported that ATMs of obese VAT profoundly expressed miR-210-3p expression (23); however, the expression pattern of miR-210-3p on other prominent cell types like pre-adipocytes or mature adipocytes in obese ATenv has not yet been explored. Comparative analysis of miR-210-3p expression demonstrated significantly higher levels in macrophages than preadipocytes and adipocytes in obese subjects and HFD mice in contrast to lean counterparts. (Fig. S1, A and B). A similar trend was observed in 3T3-L1 preadipocytes, differentiated adipocytes, and RAW264.7 macrophages when exposed to hypoxia (H, 1% O2) and lipid (L, 0.75 mM Palmitate) for 16 h to mimic the pathophysiological obese ATenv (Fig. S1C). All these observations indicate ATMs, a major component of the stromal vascular fraction, represent a key cellular source of miR-210-3p.
Figure 1.
Obese ATMs-derived extracellular vesicles bestow miR-210-3p to adipocytes.A and B, representative immunofluorescence staining of F4/80 (red), Pref1 (green) with DAPI counterstaining in the VAT (eWAT) section of Chow and HFD-fed mice followed by confocal microscopy (A) and quantification analyses (B). Scale bar represents 50 μm. C and D, uniform manifold approximation and projection (UMAP) of adipose progenitor cells, mature adipocytes, and macrophages from the reanalysis of publicly available sn-Seq data; sequenced all cells of VAT in healthy and obese individuals followed by quantification of the projected cells. E and F, uniform manifold approximation and projection (UMAP) and relative fraction of adipose progenitor cells (ASPC), mature adipocytes, and macrophages from the reanalysis of publicly available sn-Seq data; sequenced all cells of VAT in standard diet (StndD) and HFD fed mice followed by quantification of the projected cells. G, representative images showing ATM (MΦ) releases extracellular vesicles (EVs) uptake by adipocytes (Adi) in the obese ATenv. Transmission electron microscopic (TEM) images of visceral adipose tissue (VAT) section of the obese HFD mice. H and I, TEM images of EVs isolated from primary culture of mouse bone marrow–derived macrophages (BMDMs) cotreated with H + L and mean size quantification ∗p < 0.05 by Student’s t test. J, visualization of control and H + L–treated macrophage released EVs on NanoSight LM10 with size analysis. K, representative image showing Vibrant DiO staining in macrophages cocultured with 3T3-L1 adipocytes in transwell setup. Adipocytes were stained with oil red o dye. Scale bar represents 20 μm. L, representative image showing FAM-conjugated miR-210-3p mimic transfected macrophages cocultured with 3T3-L1 adipocytes in transwell setup and fluorescence imaging showed the presence of miR-210-3p-FAM to adipocyte. Adipocytes were stained with oil redo dye. Scale bar represents 50 μm. M, Western blot analysis showing CD63 expression in equal amount of EVs isolated from control and H + L–co-incubated macrophage culture supernatant, from StndD or HFD mice serum and ND or DM patients’ serum. N, miR-210-3p expression analyses in control and H + L–incubated macrophage-secreted EVs (n = 3); ∗∗p < 0.01 by Student’s t test. O, qRT-PCR analyses of miR-210-3p in EVs isolated from the primary culture of sorted F4/80+ ATMs from VAT of StndD and HFD mice (n = 4); ∗∗p < 0.01 by Student’st test. P and Q, miR-210-3p expression in the serum-containing EVs isolated from the serum of StnD and HFD mice (P) (n = 4); ∗∗p < 0.01; ND and DM individuals (Q) (n = 14–16); ∗∗∗∗p < 0.0001 by student’s t test. Data are expressed as mean ± SD; n = 3 to 16; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.0001 (Student’s t test). ATenv, adipose tissue microenvironment; ATM, adipose tissue macrophage; DM, obese diabetic; FAM, fluorescein amidite; HFD, high-fat diet; ND, non-obese non-diabetic; StndD, standard diet.
To examine the role of ATMs-derived miR-210-3p, we initially explored the EVs released by ATMs, as recent studies have suggested that ATMs-derived EVs bolster systemic inflammation and IR (18, 24). Transmission electron microscopy (TEM) imaging of VAT from HFD mice (16 weeks old) revealed a substantial presence of EVs within ATMs with some protruding out from the membrane (Fig. 1G). In addition, when bone marrow–derived macrophages (BMDMs) were exposed to hypoxia and lipid, the size of EVs (216.8 ± 54.3 nM) significantly increased compared to EVs of control untreated cells (175.1 ± 51.8 nM) (Fig. 1, H and I). The nanoparticle tracking analyses demonstrated a notable increase in both the average size and particle density of EVs isolated from HL-treated cells compared to untreated cells (Fig. 1J). To comprehend the fusion of ATMs-derived EVs with adipocytes under pathophysiological conditions, we performed in vitro coculture experiment of Vibrant DiO-labeled RAW264.7 macrophages on the upper boyden chamber (transwell 0.4 μm membrane), with differentiated Oil red-O–stained mature adipocytes in the lower chamber. Interestingly, fluorescence imaging showed notable uptake of Vibrant DiO dye by adipocytes indicating the delivery of macrophage-specific EVs (Fig. 1K). Subsequently, to investigate the transfer of miR-210-3p from macrophage EVs to adipocytes, we cocultured fluorescein amidite (FAM)-conjugated miR-210-3p mimic–transfected RAW264.7 macrophages with differentiated 3T3-L1 adipocytes. Fluorescence imaging displayed majority of mature adipocytes were FAM positive (Fig. 1L), suggesting the transfer of macrophage-derived miR-210-3p into adipocytes via EVs. We validated the isolated EVs from the cell culture supernatant, animal serum, and patient serum by using the EV-specific marker CD63 (25). We noted a high abundance of EVs in the serum of HFD mice, T2D patients, and macrophages treated with H+L (Fig. 1M). Further, we measured the miR-210-3p level in the isolated EVs from H+L-treated macrophages, normalized against U6 RNA as a housekeeping control. The data revealed approximately a 60-fold increase in the expression of miR-210-3p in EVs from H+L-treated macrophages compared to the control (Fig. 1N). Similarly, ex vivo culture of ATMs, isolated from epididymal fat pads of StndD and HFD mice, demonstrated a significant upregulation of miR-210-3p in the EVs of HFD mice (Fig. 1O). Moreover, the magnitude of miR-210-3p levels in the systemic circulation was assessed by extracting serum EVs from StndD and HFD mice as well as from lean non-diabetic individuals and obese T2D patients. We observed approximately 6.9-fold and 7.5-fold increases in miR-210-3p levels in the serum EVs of HFD mice and obese T2D patients compared to their respective controls (Fig. 1, P and Q), indicating rise of miR-210-3p–enriched EVs in systemic circulation during chronic obesity.
ATM-derived miR-210-3p-enriched EVs reduce glucose uptake in adipocytes by directly silencing GLUT4 expression
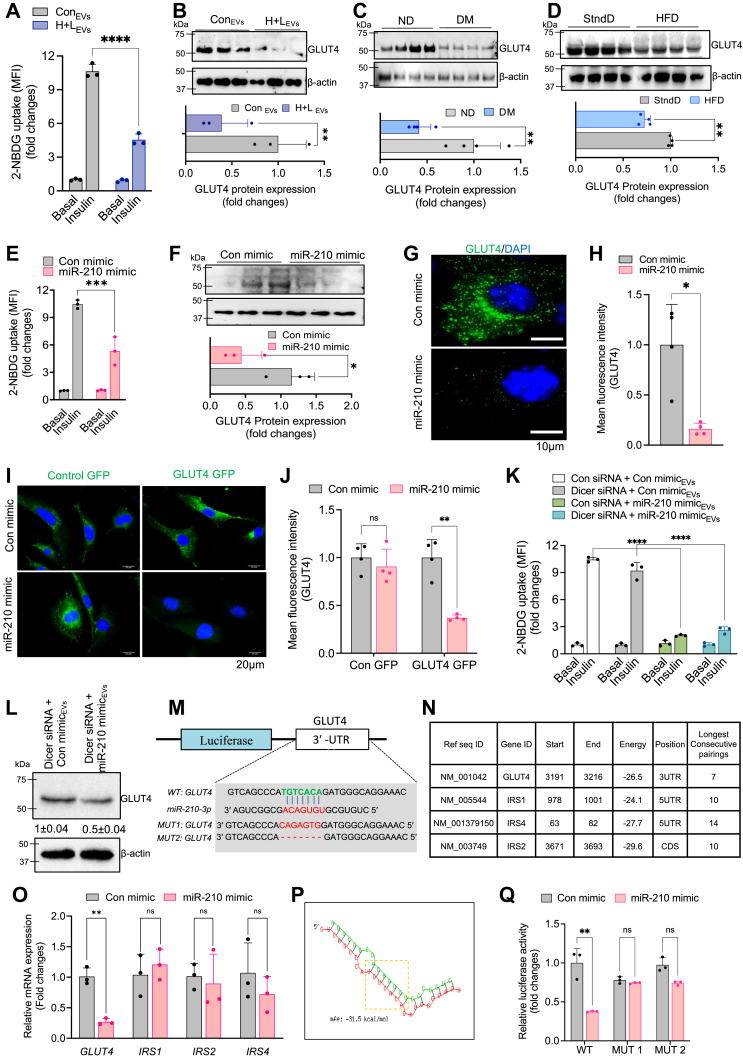
To examine the efficacy of miR-210-3p–enriched EVs on the impairment of insulin sensitivity, we treated differentiated 3T3-L1 adipocytes with EVs isolated from RAW264.7 macrophages under normoxic (con) or lipid-rich hypoxic (H + L) conditions, for 16 h. A significant reduction of glucose uptake by adipocytes was noticed in response to EVs from the H+L condition, as indicated by reduced 2-(N-(7-Nitrobenz-2-oxa-1,3-diazol-4-yl)Amino)-2-Deoxyglucose (2-NBDG) uptake (Fig. 2A). Moreover, analysis of insulin signaling pathway molecules activation revealed a considerable downregulation of GLUT4 expression in adipocytes treated with miR-210-3p–enriched EVs from the H+L condition (Fig. 2B). These findings suggest that EVs secreted from H+L-treated macrophages which encompass higher levels of miR-210-3p substantially attenuate insulin sensitivity in adipocytes. Additionally, we observed a significant reduction in GLUT4 expression in adipocytes isolated from the VAT of obese diabetic patients (Fig. 2C) and HFD mice compared to their respective controls (Fig. 2D).
Figure 2.
Loss of glycemic homeostasis associated with direct suppression of GLUT4 mediated by obese ATMs-driven miR-210-3p.A, insulin-stimulated glucose uptake was monitored by measuring 2-NBDG in the cell lysate of adipocytes, incubated with control, and H + L–treated BMDM released EVs. (n = 3); ∗∗∗∗p < 0.0001. B–D, Western blot analysis and quantification of GLUT4 protein expression in adipocytes exposed with control or H + L–incubated BMDM EVs (B); adipocytes isolated from VAT of ND and DM patients (C); adipocytes isolated from VAT of SD and HFD mice (D). E, glucose uptake in the presence or absence of insulin stimulation in control mimic or miR-210-3p mimic transfected adipocytes (n = 3); ∗∗∗p < 0.001. F, Western blot analysis showed GLUT4 expression in adipocytes transfected with control mimic and miR-210-3p mimic (n = 3), (n = 3); ∗∗p < 0.01 by Student’s t test. G and H, GLUT4 immunofluorescence imaging (G) and quantification analyses (H) in adipocytes transfected with control mimic and miR-210-3p mimic (n = 3). Scale bar represents 10 μm. ∗∗p < 0.01 by Student’s t test. I and J, immunofluorescence image showing GLUT4 migration in murine myoblast cells transfected with control mimic or miR-210-3p mimic (I) and quantification analysis (J) (n = 3); ∗∗p < 0.01, ns= not significant, by two-way ANOVA. K and L, glucose uptake assay (K) and western blotting analyses of GLUT4 (L) in adipocytes under the presence or absence of insulin stimulation in adipocytes cocultured with Dicer-silenced macrophage transfected with control mimic or miR-210-3p mimic. Quantification analyses, (n = 3), ∗∗∗∗p < 0.0001 by student’s t test. M, miRNA target prediction analyses using miRWALK database search analysis showed miR-210-3p targets for insulin signaling pathways molecule. N, mRNA expression analyses of predicted targets GLUT4, IRS1, IRS2, IRS4 in adipocytes transfected with control mimic or miR-210-3p mimic, (n = 3), ∗∗p < 0.01, ns-not significant, by student’s t test. O, the putative-binding site of GLUT4 with miR-210-3p and seed sequence of miR-210-3p and mutated 3′-UTR site of GLUT4. P, RNAhybrid webserver was used to predict the minimum free energy (mfe) of GLUT4 mRNA transcripts and miRNA-210-3p interactions. Q, luciferase activity in adipocytes transfected with WT or mutated (mut1 and mut 2) GLUT4 plasmid constructs and control mimic or miR-210-3p mimic (n = 3). ∗∗p < 0.01 by Student’s t test. Data are expressed as means ± SD; n = 3–5; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ns = non-significant (Student’s t test and two-way ANOVA). 2-NBDG, 2-(N-(7-Nitrobenz-2-oxa-1,3-diazol-4-yl)Amino)-2-Deoxyglucose; ATM, adipose tissue macrophage; BMDM, bone marrow-dervied macrophage; Con mimic, control mimic; EVs, extracellular vesicles; EV, extracellular vesicle; H, hypoxia; L, lipid; ND, non-obese non-diabetic; ns, not significant.
To further validate the role of macrophage-derived miR-210-3p–loaded EVs on adipocytes' insulin action, RAW264.7 macrophages were transfected with either miR-210-3p mimic or control mimic and co-cultured with adipocytes. We observed a significant reduction in insulin-stimulated 2-NBDG uptake and GLUT4 protein level in adipocytes when exposed to miR-210-3p mimic–transfected macrophages (Fig. 2, E and F). In addition, immunofluorescence analysis of GLUT4 protein expression in miR-210-3p mimic–transfected 3T3-L1 adipocytes demonstrated a considerable reduction in cellular GLUT4 levels (Fig. 2, G and H). Similarly, a reduction in cellular GLUT4 levels in response to miR-210-3p mimic transfection was observed in C2C12 skeletal muscle cells (Fig. 2, I and J).
Several reports have highlighted the role of EV miRNAs secreted from ATMs in the pathophysiology of metabolic diseases (18, 26, 27). Therefore, to rule out the involvement of other miRNA species in the EVs of macrophages in inducing adipocyte IR, we first silenced Dicer expression in BMDMs using siRNA, followed by the delivery of either control mimic locked nucleic acid (LNA) or miR-210-3p mimic LNA. Knockdown of Dicer expression was validated by RT-qPCR analysis (Fig. S2A). As expected, the gene expression analyses revealed a striking increase of miR-210-3p with no sign of other miRNA species like miR-27a-3p, miR-126, and miR-200b-3p in the EVs isolated from Dicer-silenced miR-210-3p mimic–transfected macrophages (Fig. S2, B and C). We treated adipocytes with those EVs isolated from miR-210-3p mimic–transfected Dicer-silenced BMDMs, where a significant reduction of 2-NBDG uptake was observed (Fig. 2K) along with the depletion of GLUT4 protein expression (Fig. 2L), indicating the potential role of macrophage EVs-associated miR-210-3p in impairing insulin-stimulated glucose uptake in adipocytes.
To investigate the direct involvement of miR-210-3p in the insulin signaling pathway, we conducted a search for putative-binding sites of miR-210-3p on the insulin signaling pathway molecules. Analysis of the miRWALK database search revealed multiple molecular targets of miR-210-3p, including GLUT4 and different isoforms of IRS (IRS1, IRS2, and IRS4) which display a critical seed sequence necessary for miR-210-3p binding (Fig. 2, M and N). We validated this finding by transfecting control mimic or miR-210-3p mimic into adipocytes. Interestingly, our findings reveal that while GLUT4 expression is significantly silenced in miR-210-3p mimic–transfected adipocytes compared to control mimic-transfected cells, there is no notable change in the expression of different IRS isoforms (Fig. 2O). This could be attributed to the position of the miR-210-3p target seed sequence in these genes, as the 3′UTR of GLUT4, CDS of IRS2, and 5′UTR of IRS1 and IRS4 contain the miR-210-3p target sequence. Additionally, we calculated the minimum free energy (−31.5 kCal/mol) required for the interaction between GLUT4 3′UTR and miR-210-3p using the RNAhybrid webserver (Fig. 2P). To confirm the binding of GLUT4 3′UTR with miR-210-3p, we performed the GLUT4 3′UTR luciferase reporter assay. WT- or mutated (MUT)-GLUT4 3′UTR luciferase plasmids were cotransfected with either control mimic or miR-210-3p mimic in 3T3-L1 adipocytes. Delivery of miR-210-3p mimic significantly repressed WT-GLUT4 3′UTR luciferase activity in comparison to control mimic-transfected cells, whereas, MUT-GLUT4 luciferase activity remained unaffected in miR-210-3p mimic–transfected cells (Fig. 2Q), indicating a direct involvement of miR-210-3p binding with GLUT4 3′UTR.
Obese ATMs-derived miR-210-3p-loaded EVs promote glucose intolerance and potentiate systemic IR
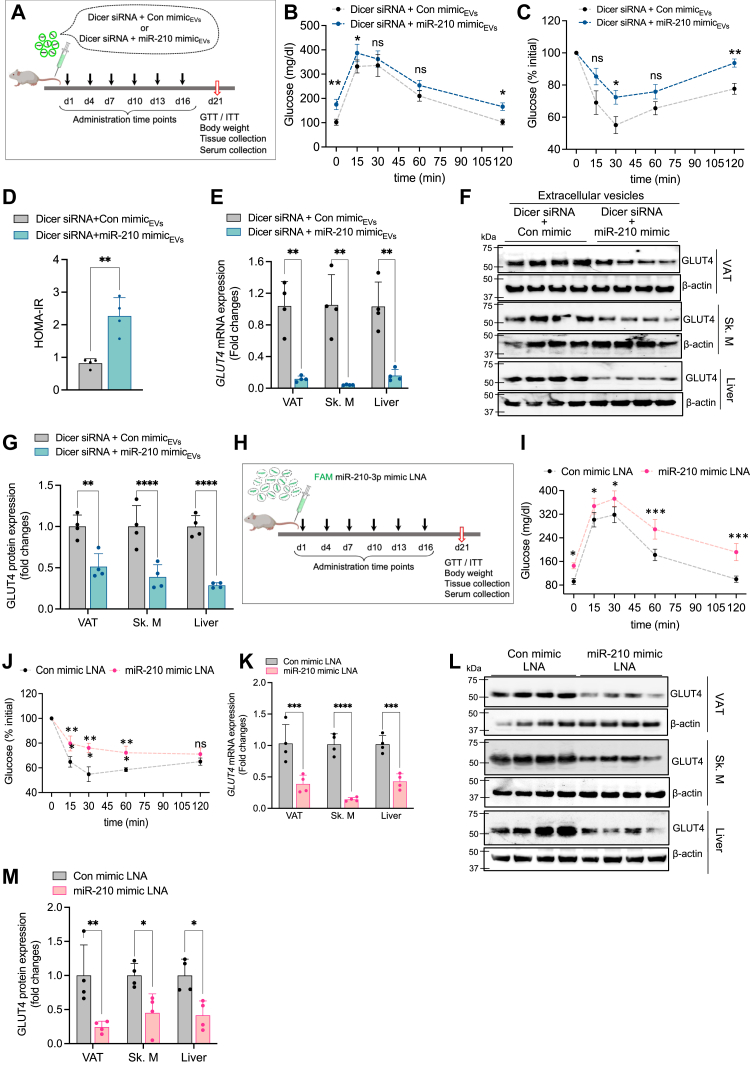
To validate our findings in vivo, we administered Vibrant DiO-labeled EVs isolated from Dicer-silenced BMDMs transfected with either control or miR-210-3p mimic LNAs to C57BL/6 lean mice via intravenous injection (2 × 106 particles/mouse) once every 3 days for 20 days (Fig. 3A). A considerable accumulation of Vibrant DiO-labeled EVs (Fig. S3A) along with a profound increase of miR-210-3p levels (Fig. S3B) was noticed in the VAT, skeletal muscle, and liver, without any significant changes in body mass (Fig. S3C). Moreover, mice administered with labeled EVs exhibited impaired glucose- and insulin-tolerance as indicated by glucose tolerance test (GTT) and insulin tolerance test (ITT) analyses (Fig. 3, B and C). Furthermore, homeostatic model assessment for insulin resistance (HOMA-IR) analysis demonstrated a substantial induction of IR (Fig. 3D). Administration of these EVs in lean mice resulted in adipocyte hypertrophy (Fig. S3D) along with a notable decrease in GLUT4 gene and protein expression in the VAT, skeletal muscle, and liver (Figs. 3, E–G and S3E). Taken together, these results highlight GLUT4 as a key molecular target of miR-210-3p affecting insulin sensitivity. All these results indicate that in vivo delivery of miR-210-3p–loaded EVs promotes glucose intolerance and IR in mice through the paracrine and endocrine actions on the VAT, skeletal muscle, and liver. For direct evidence, we administered FAM-conjugated control or miR-210-3p mimic LNAs loaded in invivofectamine, to lean mice via intravenous injections six times over 21 days at regular intervals (Fig. 3H). Fluorescence imaging and RT-qPCR analysis revealed increased localization and abundance of miR-210-3p in the VAT, skeletal muscle, and liver of mice administered with labeled miR-210-3p LNA with no change in body weight (Fig. S3, F–H). These mice exhibited disrupted systemic glucose homeostasis and insulin responsiveness (Fig. 3, I and J).
Figure 3.
miR-210-3p–enriched EVs of obese ATMs promote glucose intolerance and insulin resistance in vivo.A, schematic diagram showing standard diet–fed C57BL/6 mice administered with EVs isolated from Dicer-silenced macrophages transfected with control mimic and miR-210-3p mimic via an intravenous route with a regular interval for 21 days before termination of the experiment. B–D, glucose tolerance test (GTT) (B), insulin tolerance test (ITT) (C), and HOMA-IR (D) analyses were performed in StndD fed–mice injected with EVs isolated from Dicer-silenced macrophage transfected with control mimic LNA or miR-210-3p mimic LNA (n = 4). ∗p < 0.05, ∗∗p < 0.01, ns-non-significant by two-way ANOVA and ∗∗p < 0.01 unpaired Student’s t test. E–G, GLUT4 mRNA expression analyses (E), western blotting (F), and quantification analyses (G) in visceral adipose tissue (AT), Liver, and skeletal muscle (SM) in standard diet–fed C57BL/6 mice administered with Dicer-silenced macrophage exosomes having control mimic LNA or miR-210-3p mimic LNA via the intravenous route in regular intervals for 21 days (n = 4), ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 by two-way ANOVA. H, schematic diagram showing control mimic or miR-210-3p mimic LNA intravenous injection in regular intervals for 21 days in C57BL/6 mice. I and J, glucose tolerance test (I) and insulin tolerance test (J) were performed in standard diet–fed mice administered with control mimic LNA and miR-210-3p mimic LNA with each 3 days interval in the period of 21 days, (n = 4), ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ns non-significant by two-way ANOVA. K–M, qRT-PCR analyses (K), immunoblotting (L), and quantification analyses (M) of GLUT4, in standard diet–fed C57BL/6 mice injected with control mimic LNA or miR-210-3p mimic LNA via the intravenous route in regular intervals for 21 days, (n = 4), ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 two-way ANOVA. Data are expressed as means ± SD; n = 3 to 4. ATM, adipose tissue macrophage; EV, extracellular vesicle; HOMA-IR, homeostatic model assessment for insulin resistance; LNA, locked nucleic acid; StndD, standard diet.
Further, we evaluate the efficacy of miR-210-3p mimic LNA administration on GLUT4 expression in insulin target tissue of C57BL/6 lean mice. A striking reduction of GLUT4 mRNA (Fig. 3K) and protein (Fig. 3, L and M) expressions were noticed in the VAT, skeletal muscle, and liver of mice administered with miR-210-3p mimic LNA. Interestingly, H&E staining and GLUT4 immunostaining of the VAT displayed enlarged adipocytes which coincided with subdued expression of GLUT4 in miR-210-3p mimic LNA-administered mice (Fig. S3, I and J). Collectively, these findings indicate that macrophage EVs-derived miR-210-3p localizes to insulin target tissues and contributes to systemic glucose intolerance and IR.
Targeted inhibition of miR-210-3p in VAT rescue obese HFD mice from glucose intolerance and IR
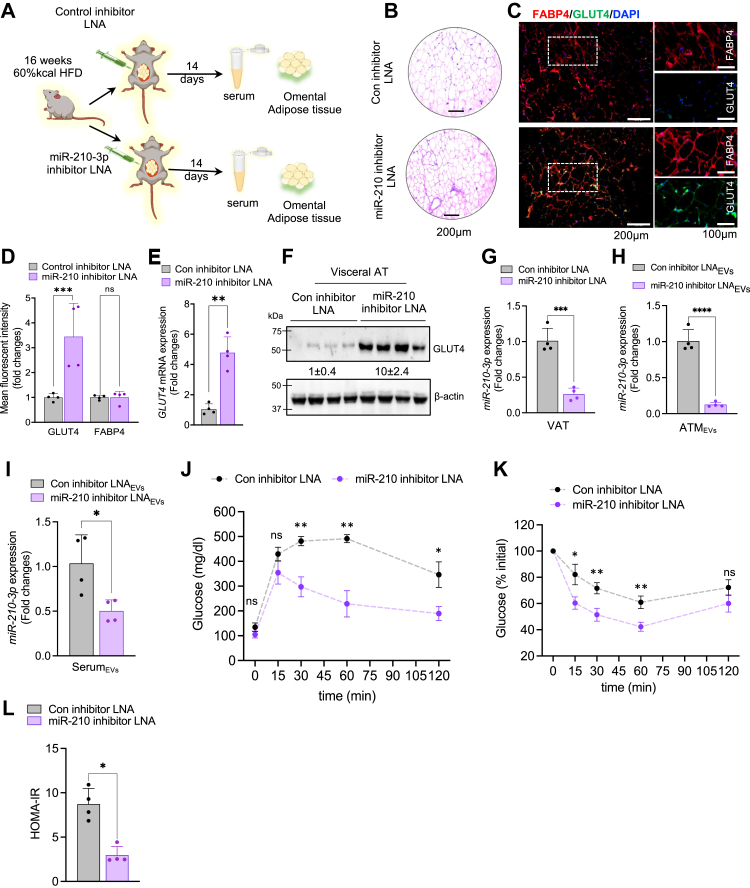
To examine the therapeutic potential of anti-miR-210-3p in alleviating obesity-induced impairment of IR, we delivered invivofectamine-encapsulated FAM-labeled control inhibitor LNAs or miR-210-3p inhibitor LNAs directly to the VAT of HFD-fed obese mice for a period of 14 days followed by the collection of serum and tissue samples (Fig. 4A). The HFD mice exhibited a substantial decrease in GLUT4 expression in the VAT, skeletal muscle, and liver, compared to StndD mice (Fig. S4, A–C). The presence of FAM-labeled miR-210-3p inhibitor LNA was confirmed in the VAT, skeletal muscle, and liver (Fig. S4D) which correlated with reduced adipocyte hypertrophy in the VAT of miR-210-3p inhibitor LNA-administered HFD mice (Fig. 4B). Administration of miR-210-3p inhibitor LNAs in the VAT of HFD mice led to a notable increase in GLUT4 levels in the VAT (Fig. 4, C–F), skeletal muscle (Fig. S4, E and F), and liver (Fig. S4, G and H), which coincided with a subdued level of miR-210-3p in the VAT, EVs released by ATM, and in the serum EVs (Fig. 4, G–I). VAT-specific delivery of miR-210-3p inhibitor LNA-enriched EVs significantly improved systemic glucose tolerance and insulin sensitivity in HFD mice as evidenced by GTT, ITT, and HOMA-IR analyses (Fig. 4, J–L). Overall, our findings highlight miR-210-3p as a promising therapeutic candidate for addressing obesity-induced IR and T2D.
Figure 4.
miR-210-3p inhibition rescue DIO mice from glucose intolerance and insulin resistance.A, schematic diagram showing control inhibitor LNA or miR-210 inhibitor LNA administration in the visceral adipose tissue of 12 weeks HFD mice. B, H&E staining of VAT section of control inhibitor LNA or miR-210 inhibitor LNA-treated HFD mice. Scale bar represents 200 μm. C and D, immunofluorescence imaging (C) and fluorescent intensity quantification analyses (D) of GLUT4 (green), and FABP4 (red) in the VAT section of control inhibitor LNA and miR-210 inhibitor administered HFD mice, (n = 4), ∗p < 0.05, ∗∗∗p < 0.001, ns = not significant. Scale bar represents 200 μm. E and F, GLUT4 mRNA expression analyses (E) and western blotting (F) analyses in the visceral adipose tissue (VAT) of control inhibitor LNA or miR-210 inhibitor LNA-treated HFD mice, (n = 4), ∗∗p < 0.01 by Student’s t test. G, miR-210-3p expression in the visceral adipose tissue of control inhibitor LNA or miR-210 inhibitor LNA mice (n = 4). H, miR-210-3p expression in the ATM-derived EVs isolated from the VAT of control inhibitor or miR-210-3p inhibitor LNA–administered HFD mice (n = 4), ∗∗∗∗p < 0.0001 by Student’s t test. I, miR-210-3p expression in the EVs isolated from the serum of control inhibitor LNA and miR-210-3p inhibitor LNA–administered HFD mice, (n = 4), ∗p < 0.05 by Student’s t test. J–L, glucose tolerance test (GTT) (J), insulin tolerance test (ITT) (K), and HOMA of insulin resistance (HOMA-IR) (L) analyses were performed in HFD mice injected with control inhibitor LNA or miR-210 inhibitor LNA (n = 4). ∗p < 0.05, ∗∗p < 0.01, ns= not significant by two-way ANOVA and ∗p < 0.05 by student’s t test. Data are expressed as means ± SD. ATM, adipose tissue macrophage; EV, extracellular vesicle; HOMA-IR, homeostatic model assessment for insulin resistance; LNA, locked nucleic acid.
Discussion
Obesity-induced pathophysiological ATenv allows massive ATM infiltration and alteration of their polarity towards a proinflammatory state contributing to chronic AT inflammation and insulin resistance (21, 23). It has been shown that the alteration of the miRNA profile in the adipose tissue play a critical role in modulating insulin signaling pathway during obesity. Reportedly, miR-128, miR-103, miR-107, and miR-26b were identified as altered miRNAs in adipocytes, influencing insulin sensitivity (28, 29, 30). Of these, miR-128 increases specifically in adipocytes under hypoxic conditions which attenuates insulin sensitivity by suppressing insulin receptor expression (28). However, these studies primarily focused on adipocyte-specific miRNAs and their effect on insulin sensitivity without considering (i) other environmental factors like lipid content and hypoxia, (ii) diverse cell sources such as ATMs or other stromal cell populations, and (iii) their active participation in systemic insulin resistance.
miRNAs have the potential to target multiple mRNA molecules, while several miRNAs can target a single mRNA species (31). Additionally, miRNAs are often packaged and transported to distant organs via EVs and can modulate the function of other cell types (13, 16, 32, 33). Macrophages, the most abundant stromal cells in adipose tissue, are known to secrete varied sizes of EVs, including exomeres, exosomes, and microvesicles that contribute to adipose tissue dysfunction and inflammation (18, 26, 34, 35). For instance, microvesicles released from M1-type pro-inflammatory macrophages induce insulin resistance in primary human adipocytes by activating the NF-κB signaling pathway and downregulating GLUT4 translocation (36). Recent studies have shown that lean insulin-sensitive mice readily develop systemic insulin resistance and glucose intolerance when exposed to exosomes (Exos) derived from the ATMs of obese mice, whereas an inverse effect was observed in obese insulin-resistant mice exhibiting improved insulin sensitivity when administered with ATMs-derived Exos from lean insulin-sensitive mice (18, 37). It has now been widely recognized that a substantial disparity in the quantity of differentially expressed miRNAs between the ATMs and ATMs-derived exosomes from lean and obese mice/subjects (38, 39). Analysis of ATMs-derived Exos from obese mice revealed significant upregulation of miR-155, a major contributor to glucose intolerance by suppressing PPARγ (18). However, despite these findings, the study did not pay attention to adipose tissue environmental cues like hypoxia; thus, we are interested to know about lipid-rich hypoxic ATenv-induced ATM-specific hypoxamiRs and their roles in insulin resistance during obesity. To date, hypoxamiRs that could directly target insulin signaling pathway molecules have not been revealed. In our previous study, we identified that miR-210-3p, out of many hypoxamiRs, has been greatly increased in the ATMs of obese mice, playing a crucial role in inflammation by targeting the SOCS1–NF-κB pathway (23). The present study delves into how the lipid-enriched hypoxic ATenv in obesity drives the pathogenesis of insulin resistance and glucose intolerance through the release and delivery of miR-210-3p–loaded EVs from ATMs to insulin target cells such as adipocytes, skeletal muscle cells, and hepatocytes via paracrine and endocrine actions. EVs isolated from the ATMs of obese T2D patients and HFD mice are highly enriched with miR-210-3p compared to the lean nondiabetic subjects or StndD mice.
To examine the exclusive role of ATMs-derived miR-210-3p–loaded EVs in disrupting insulin signaling pathway, we have performed various experiments that evidenced that EVs isolated from miR-210-3p–transfected Dicer-silenced BMDMs promote glucose intolerance and insulin insensitivity in lean mice by reducing GLUT4 expression in the VAT, skeletal muscle, and liver. This experimental design was influenced by the previous study of AT-derived circulatory miRNAs using AT-specific Dicer KO mice, which exhibited dysfunction in miRNA processing and a reduction of AT-specific overexpressed miRNAs such as miR-201, miR-222, miR-221, miR-16 (39). This article established the role of adipose tissue as a major repertoire for EVs-enriched miRNAs found in circulation. Our findings, obtained by administering miR-210-3p–enriched EVs from the Dicer-silenced BMDM to the lean mice, nullify the involvement of any other miRNAs in ATMs-derived EVs in obesity-induced insulin resistance. However, to confirm miR-210-3p′s role in regulating cellular insulin sensitivity, we monitored glucose uptake in adipocytes transfected with the miR-210-3p mimics. A notable reduction in insulin-stimulated glucose uptake was noticed in adipocytes post miR-210-3p mimic transfection, clarifying the direct participation of miR-210-3p on insulin resistance. Bioinformatic analyses revealed multiple molecules, including GLUT4, in the insulin signaling pathway that could be targeted by miR-210-3p. The 3T3L1 adipocytes transfected with miR-210-3p mimic exhibited reduced expression and translocation of GLUT4, an essential molecule within the insulin signaling pathway (40), suggesting a possible direct targeting of GLUT4 by miR-210-3p. Due to miR-210-3p′s heightened efficacy in suppressing the GLUT4 molecule, we conducted a luciferase assay, which demonstrated direct binding of miR-210-3p to the 3′UTR of GLUT4 mRNA. This binding resulted in the suppression of GLUT4 expression in adipocytes, consequently reducing cellular insulin responsiveness and glucose uptake. Moreover, delivery of invivofectamine-encapsulated miR-210-3p LNA to lean mice notably exacerbates metabolic dysfunction as indicated by the impaired glucose- and insulin-tolerance test, which, coupled with the significant reduction in GLUT4 expression in the VAT, skeletal muscle, and liver. Hence, our study unveils a novel mechanism in which EVs carrying miR-210-3p from obese ATMs contribute to systemic insulin resistance, potentially linked to glucose intolerance.
Further, the intervention study assessed the therapeutic potential of anti-miR-210-3p LNA by delivering it to the VAT of HFD mice. Surprisingly, we discovered the anti-miR-210-3p administration conducive to the amelioration of metabolic health with improved glucose tolerance and insulin sensitivity. Mice treated with anti-miR-210-3p showed increased expression of GLUT4 in the VAT, liver, and skeletal muscle. Thus, inhibiting miR-210-3p in the VAT could serve as a promising therapeutic strategy to combat obesity-induced systemic glucose intolerance and insulin resistance. However, further studies on ATM-specific miR-210-3p–deficient transgenic animal models will provide direct evidence of miR-210-3p role in the understanding of disease progression, which is a potential limitation of our study.
In summary, our findings demonstrated that miR-210-3p–enriched EVs secreted from the ATMs of obese VAT travel to adipocytes, skeletal muscle, and liver via paracrine and endocrine actions and induces systemic IR and glucose intolerance by directly targeting GLUT4, a key molecule in the insulin signaling cascade. In contrast, the delivery of anti-miR-210-3p LNA rescues obese HFD mice from the pathophysiological state of glucose intolerance by improving insulin sensitivity in target tissues. Thus, our study uncovers the direct role of ATM-derived miR-210-3p–enriched EVs in causing systemic IR and could serve as a potential therapeutic target for managing T2D.
Experimental procedures
Primary cells
For mouse BMDMs, we surgically removed the femur and tibia from 8 to 10 weeks C57BL/6J mice in a sterile environment, and flushed out bone marrow cells, and differentiated into BMDM following literature 20. Briefly, epiphyses from bone marrow were removed and were gently washed with 2 ml of PBS, 2 to 3 times. The bone marrow cells were collected in a 50 ml tube followed by centrifuge at 200g for 5 min at 4 °C. Cell pellets were resuspended with chilled RBC lysis solution for 5 min followed by centrifuge at 200g for 5 min at 4 °C. The BM cell pellet was further resuspended in a growth media (Dulbecco’s modified Eagle’s medium (DMEM) high glucose with 10% HI-FBS, 1% Pen-Strep) containing 1 ng/ml mouse M-CSF1 recombinant protein and passed through a 100 μM sterile cell strainer to remove debris and macroparticles. Cells were seeded in a 6-well plate with 0.5 × 106 cells per well and 3 × 106 cells in a 100 mm dish, cultured for 6 days before further experiments.
We cultured ATMs (F4/80+), sorted from the stromal vascular fraction, which was isolated from the visceral adipose tissue of lean and obese mice following Collagenase I digestion. The sorting of macrophages was performed in BD FACS Aria Cell Sorter (Franklin Lakes). The macrophages were cultured in RPMI 1640 supplemented with 10% fetal bovine serum (FBS) and 1% Penicillin-Streptomycin Solution at 37 °C in a humidified atmosphere with 5% CO2 before isolation of EVs released from these cells. All treatments were given in serum and antibiotics-free media, and EVs isolation from these cultures was performed in cells having fresh serum and antibiotics-free media, changed at least 16 h before termination of the experiment.
Cell lines
3T3-L1 cells were cultured in a growth medium, containing DMEM supplemented with 10% bovine calf serum and 1% Penicillin-Streptomycin Solution. 3T3-L1 preadipocytes were differentiated into adipocytes following chemically induced differentiation protocol of ATCC, such as medium containing DMEM with high glucose, HI-FBS, 1.0 μM dexamethasone, 0.5 mM IBMX, and 1.0 μg/ml insulin were added upon full confluency. Later on, the differentiation medium was replaced by adipocyte maintenance medium containing DMEM with high glucose, 10% HI-FBS, and 1.0 μg/ml insulin. C2C12 myoblast cells were cultured in DMEM, supplemented with 10% HI-FBS and 1% Penicillin-Streptomycin Solution. RAW264.7 macrophage cells were cultured in DMEM supplemented with 10% FBS and 1% Penicillin-Streptomycin Solution at 37 °C in a humidified atmosphere with 5% CO2. We incubated RAW264.7 macrophages with a fixed concentration of palmitate (0.75 mM) and exposed them to hypoxia condition (1% O2 and 5% CO2) for different time periods in the Heracell VIOS 160i incubator.
Animals
WT C57BL/6J male mice aged 4 to 5 weeks and weighed 18 to 22 g were kept in the NIPER Mohali animal house facility for 5 to 6 days in 12 light/dark cycle at 23 °C ± 2 deg. C with relative humidity 55 ± 5% and fed with normal pellet diet and water ad libitum. For the development of a diet-induced obese and insulin resistance model, C57BL/6J mice were fed with HFD pellets having 60% kcal of fat for 12 weeks. All other mice were fed with provide StndD having 10% kcal of fat for 12 weeks. All experimental animals have free access to sterilized water and food. The blood glucose level has been measured regularly with Accu-Chek glucometer (Roche). Mice fed with HFD diet for 12 weeks were considered for anti-miR-210-3p LNA delivery. Briefly, mice were anesthetized by low-dose isoflurane inhalation as per standard recommendations. We then created a small incision on the abdominal site and took out the epididymal fat pads from the abdominal cavity. A total of 100 nMmiRCURY LNA miRNA power inhibitor/(anti)mmu-miR-210-3p or miRCURY LNA control inhibitor was injected in ten different sites on both sides of the abdominal visceral WAT. The skin layer was stitched carefully using Ethicon absorbable surgical suture. On day 14 of post-surgery, animals were utilized for different experiments.
We have evaluated the role of miR-210-3p on the development of insulin resistance by administering nanoparticle-encapsulated control mimic or FAM-conjugated miR-210-3p mimic (2.5 mg/kg bw) in the tail-vein of lean StndD-fed mice for 6 times in the span of 21 days. The dose for each injection was 5 nmol of mimic/mice. In the termination of experiments, we harvested epididymal fat pads, Liver, and skeletal muscle to visualize the presence of fluorescent-conjugated mimic.
To decipher the role, miR-210-3p–loaded EVs of 8 weeks aged C57BL/6J mice fed with StndD were intravenously administered with BMDMs-derived extracellular vesicles (2 × 106 EVs per animal) particles with an interval of 3 days for 21 days (total 6 times). To isolate EVs, BMDMs were co-transfected with Dicer siRNA and control mimic or Dicer siRNA and miR-210-3p mimic followed by the collection of conditioned media and EVs isolation. To monitor the in vivo trafficking of EVs, BMDMs were stained with Vibrant DiO (Invitrogen) before EVs isolation. Fluorescent-labeled EVs were isolated, purified, and administered through tail-vein injection in mice. The confirmation was done by taking out biopsies of VAT, Liver, and skeletal muscles.
We measured the body weight of the animals regularly and determined the GTT by measuring blood glucose levels before and after oral gavages of 1 g glucose/kg bw at the indicated time points before the termination of all of the animal experiments. Similarly, ITT was performed by injecting 1 IU insulin/kg bw. We have used the following formula to calculate the HOMA-IR: fasting insulin level (microU/L) × fasting glucose level (nmol/l)/22.5 (41). For histochemistry analyses, a small portion of harvested tissues were freshly frozen followed by cryosectioning.
All animal experiments were performed following the guidelines prescribed by and with the approval of the Institutional Animal Ethics Committee NIPER Mohali, Punjab.
Human participants
A total of 13 men and 17 women have participated in this study. The study population was categorized into two groups based on BMI and blood glucose level. Study subjects having BMI 18 to 25 kg m–2 with fasting blood glucose level <85 were considered as lean non-diabetic group (n = 16) whereas patients with BMI >30 kg m–2 and fasting blood glucose level >120 were considered as an obese diabetic group (n = 14) as presented in the Table S2. In this study, surgically dissected visceral adipose tissue samples and blood samples were collected from the patients who were admitted to the Dayanand Medical College & Hospital, Ludhiana, Punjab and underwent abdominal surgery. The study protocol for the use of human blood and tissue samples was approved by the Institute Ethics Committee, Dayanand Medical College & Hospital, Ludhiana, Punjab. We have obtained written informed consent from all participants in this study. The human study described in our manuscript adheres to the principles outlined in the Declaration of Helsinki.
Extracellular vesicle experiments
Isolation and characterization
The macrophage (BMDMs or RAW264.7) cultured media was collected for EV isolation. After 48 h of transfection experiments, the complete media was replaced with serum and antibiotics-free media and kept for 16 h for EVs isolation using EV isolation kit. Prior to initiating the isolation process, the conditioned media were centrifuged at 3000g for 15 min and the supernatant was transferred to a fresh tube to remove cells and debris. We have isolated EVs from serum samples of both humans and mice using the ExoQuick ULTRA kit following manufacturer’s protocol.
Nanoparticle tracking analyses
After isolation and purification of EVs, it was subjected to concentration and size analyses using Nanosight LM10 NTA analyses these vesicles using (Malvern Instruments).
Transmission Electron Microscopy
We measured the size of EVs by performing TEM. Briefly, the purified EVs were fixed using 2% paraformaldehyde for 5 min at room temperature. Five to seven microliters of exosomal suspension was loaded onto the carbon-coated copper grid and incubated for 1 min, followed by staining using 2% of uranyl acetate solution on the surface of the EM grid by syringe. The excess staining solution was removed using filter paper & rinsed the grid with a drop of water & the grid was subjected to drying for 30 min & then examined by using TEM (Model TF-20), FEI, at NIPER, S.A.S Nagar, Punjab.
RNA and protein isolation from EVs
EVs isolated from cell culture media or serum with a particle number of 3 × 106 – 5 × 106 were considered for miRNA isolation and 1 × 107 – 1 × 108 EVs were considered for protein isolation using total exosome RNA and protein isolation kit following manufacturer protocol.
TEM of tissue sections
The experiment was performed following protocol reported earlier (42). Briefly, a small portion of adipose tissue was subjected to fixation using 2.5% phosphate-buffered glutaraldehyde for at least 1 h at room temperature, followed by post-fixation using 1% osmium tetraoxide for 1 h. The samples are subsequently washed using PBS to remove the traces of osmium tetraoxide. The samples were dehydrated in ethanol, followed by propylene oxide, and then embedded in epoxy resin. The samples were sectioned using the glass knife and contrast stained using 4% uranyl acetate. These thin sections were placed on the copper grid & then were examined by using TEM (Model TF-20), FEI, at NIPER, S.A.S Nagar, Punjab, India.
miR-210-3p mimic transfection
For transfection of miR-210-3p mimic and control mimic, LipofectamineRNAiMAX transfection reagent was used according to the manufacturer’s protocol. Briefly, BMDM, RAW264.7 macrophages, or 3T3-L1 adipocytes(differentiated) were cultured in a 6-well plate in an antibiotic-free complete growth medium prior to transfection. For each well, 50 nM of miR-210-3p mimic/control mimic in Lipofectamine RNAi MAX reagent was added separately into the OptiMEM serum-free medium. Both these solutions were mixed and incubated for 5 min. The transfection mixture was added to the cells containing complete growth medium and incubated for 48 h. After 48 h of transfection, cells were washed; a fresh complete growth medium was added and used for different treatments.
Transwell co-culture experiment
RAW264.7 macrophages (1 × 105 cells/well) were cultured on transwell cell culture insert for six well plates (0.4-μm pore size, Corning) transfected with control mimic or FAM-conjugated miR-210-3p mimic kept for 24 h. The media was changed with fresh complete growth media, followed by transferring the insert into a well of six well plates having mature adipocytes and continued for another 24 h prior to terminating or setting up an experiment. Macrophages were seeded on the trans-well culture insert and stained with Vibrant DiO for 24 h followed by changing media and transfer to a new well having mature adipocytes. The cells in this co-culture setup have been co-exposed to a hypoxic environment along with lipid induction for 16 h and were considered for microscopy.
Glucose uptake assay
3T3-L1 adipocytes and C2C12 cells were cultured and transfected with miR-210-3p mimic or control mimic and kept on the same media for 48 h. Then cells were washed several times with PBS and serum-starved in Kreb’s Ringer Bicarbonate Buffer supplemented with 0.2% bovine serum albumin (BSA) for 6–12 h. Upon completion of starvation, these cells were utilized for glucose uptake assay using a Glucose Uptake Cell-Based Assay Kit following the manufacturer’s instruction. Briefly, insulin (100 nM) was added to the control and treated adipocytes and incubated for 30 min. Fluorescent-labeled glucose analog 2-NBDG was added to each of the incubations for 10 min before termination of the experiment. Cells were then lysed and fluorescent intensity was measured by a Microplate Reader (BMG Labtech Allmendgrün 8, 77799).
Cloning of GLUT4 3′UTR
The GLUT4 3′UTR region was PCR amplified from the genomic DNA of human PBMC using specific cloning primers. The cloning primers were designed using the Takara Bio cloning primer design tool (https://www.takarabio.com/learning-centers/cloning/primer-design-and-other-tools) for targeted amplification of GLUT4 3′UTR region flanked by XbaI and SalI restriction sites. The forward primer contained the XbaI site and the reverse primer contained the SalI site and the sequence details are provided in Table S1. The PCR amplification of the GLUT4 3′UTR region was performed with a melting temperature (Tm) of 70 °C. After 35 cycles of PCR reaction, we confirmed the presence of a band of approximately 1.35 kb in agarose gel electrophoresis that specifies targeted amplification of GLUT4 3′UTR region. The target band representing the GLUT4 3′UTR region was subjected to gel purification using the GeneJET Gel Extraction Kit following the manufacturer's instructions. For the construction of the GLUT4 3′UTR-luciferase plasmid, we used pGL3-Myb-3′UTR-luciferase vector (cat. no. #25798, Addgene) (43) as the backbone, which pGL3-MYB-3′UTR was a gift from Judy Lieberman (Addgene plasmid # 25798; http://n2t.net/addgene:25798; RRID: Addgene_25798). We performed restriction digestion of the pGL3-Myb-3′UTR-luciferase vector using XbaI and SalI enzymes for the removal of the Myb-3′UTR region from the vector. For this, 1 μg plasmid was digested by 20 units of each restriction enzyme and incubated for 45 min at room temperature followed by the heat-inactivation at 65 °C for 15 min. The digested reaction mixture was run on 0.8% agarose gel electrophoresis and the target band representing pGL3-luciferase plasmid was subjected to gel extraction. The concentration of the purified plasmid was measured and used for the ligation with purified GLUT4 3′UTR region flanked by XbaI and SalI sites for creating GLUT4 3′UTR-luciferase plasmid. For ligation, 100 ng of the purified pGL3-luciferase plasmid was incubated with 74.2 ng of the purified GLUT4 3′UTR region in the presence of T4 DNA ligase at room temperature for 15 min following manufacturers’ protocol. The ratio of insert to vector DNA was maintained at 3:1, as calculated using the NEBio Calculator. Subsequently, the ligation mixture was transformed into DH5α competent cells and plated on agar plates containing ampicillin (50 μg/ml). After transformation, single colonies were picked and inoculated in LB Broth for 6 to 8 h at 37 °C for plasmid amplification. Plasmids were then isolated for further experiments.
Site-directed mutagenesis
WT GLUT4 3′-UTR plasmid construct was used as template for the generation of mutated GLUT4 3′-UTR plasmids by using QuickChange Lightning Multi Site-Directed Mutagenesis Kit following manufacture’s protocol. Primers used to generate the mutated GLUT4 3′-UTR plasmids were designed with the help of QuikChange Primer Design Program available online at www.agilent.com/genomics/qcpd. Primer sequences used for mutated GLUT41 3′-UTR plasmids construction are listed in Table S1.
GLUT4 3′-UTR luciferase reporter assay
3T3-L1 adipocytes were co-transfected with 500 ng of WT or mutated GLUT4 3′-UTR plasmid and with either control mimic or miR-210-3p mimic (Dharmacon) using Lipofectamine LTX/Plus Reagent for 48 h in a 24-well plate. Upon termination of treatment, adipocytes were lysed, and luciferase activity was determined using Luciferase Reporter Assay System in GloMax Navigator Microplate Luminometer (Promega) following the manufacturer’s protocol.
Immunocytochemistry
3T3-L1 cells were grown on a sterile glass coverslip and differentiated into adipocytes prior to transfecting with control mimic and miR-210-3p mimic for 48 h. Adipocytes were washed with PBS and fixed with ice-cold methanol for 5 min. For intracellular staining, cells were permeabilized with 0.1% TritonX-100 in PBS for 10 min at room temperature. Cells were blocked with 1% BSA in PBS containing 0.025% Tween-20 for 30 min at room temperature and incubated with primary antibodies for 1 h at room temperature. Cells were then washed with ice-cold PBS thrice for 5 min each, followed by the incubation with fluorescence-conjugated secondary antibodies for 1 h at room temperature in the dark. Before mounting on a glass slide, cells were washed thrice for 5 min each with ice-cold PBS. Coverslips were mounted onto glass slides using anti-fade mounting medium with DAPI. Cellular images were captured by an inverted fluorescent microscope (Leica DMi8) and image analysis was performed using LAS X software (www.leica-microsystems.com/products/microscope-software/p/leica-las-x-ls/). Fluorescence intensity was quantified using ImageJ software (1.48v, NIH, USA).
Oil-red O staining
The differentiation of 3T3-L1 cells into adipocytes was further confirmed by performing oil-red O staining and imaging following the protocol described earlier (44). Briefly, adipocytes were fixed in a solution containing 4% formaldehyde for a duration of 10 min. After fixation, the cells were stained with Oil-Red O stain for 30 min at room temperature. Subsequently, the cells were rinsed with 60% isopropanol followed by three washes with PBS. To capture cellular images, an inverted fluorescent microscope (Leica DMi8) was used.
Immunostaining and confocal microscopy
Adipose tissue samples collected from human subjects and mice models were immediately washed in sterile saline and then placed in Neutral buffer formalin (10%) for overnight fixation at 4 °C. After fixation, adipose tissues were passed through increasing concentration of sucrose solution (10%, 15%, 20%) followed by embedded in optimal cutting temperature compound (Sigma) and frozen at −60 °C followed by cryosections using Cryotome with internal temperature less than −25 °C (Leica CM 1860, Leica Biosystem). Immunostaining was performed on tissue cryosections using specific antibodies. Briefly, tissue cryosections (10 μm) were placed in gelatin-coated glass slides, fixed in ice-cold methanol for 5 min, blocked with 5% BSA-containing blocking buffer, and incubated with specific primary antibodies for 1 h at room temperature. After washing, signal was visualized by subsequent incubation with fluorescence-conjugated appropriate secondary antibodies and counter-stained with anti-fade mounting medium containing DAPI. Images were captured by a Confocal microscope (LSM 880 Carl Zeiss) and analysis was performed using Zen software (https://www.micro-shop.zeiss.com/en/de/softwarefinder/software-categories/zen-black/zen-black-system/).
H&E staining and imaging
The visceral adipose tissue from animals was collected and subjected to histopathological analysis. Cryo-sectioning of the tissues was performed, as previously described in this manuscript. These tissue sections were placed on gelatin-coated glass slides and subjected to regressive staining using the following steps: 100% alcohol was passed over the sections for 20 s, repeated twice; followed by 90% alcohol for 20 s, repeated twice; 80% alcohol for 20 s; 70% alcohol for 20 s; 50% alcohol for 20 s. Subsequently, the slides were rinsed with dH2O for 1 min. Next, the slides were incubated in Hematoxylin for 3 min, followed by a 2-min water rinse. The slides were then dipped three times in a 0.3% acetic acid–containing alcohol solution, rinsed in dH2O, and further dipped several times in 0.3% ammonium water, inducing bluing upon rinsing in water. Afterward, the slides were passed through 80% alcohol for 20 s, followed by staining with 2% eosin for 30 s. To remove excess staining, the sections were washed with 95% alcohol for 20 s, repeated twice, and then with 100% alcohol for 20 s. Finally, the slides underwent a few brief dips in xylene before mounting using DPX solution. Subsequently, the H&E-stained slides were examined using a Leica DMi8 microscope for imaging.
RNA extraction and quantitative PCR
Total RNA was extracted from the cells and tissue was performed by adding TRIzol (Invitrogen) following the protocol described earlier (45). Hundred to two hundred milligrams of tissue was lysed using Tissue LyserII (Qiagen) in TRIzol solution. RNA concentration was quantified using NanoDropOneC spectrophotometer (Thermo Fisher Scientific). RNA (100 ng) was then treated with DNase I and reverse transcribed using the iScript cDNA Synthesis Kit. We used PowerUp SYBR Green Master Mix to perform real time quantitative PCR in QuantStudio3 Real-Time PCR System (Applied Biosystems) using gene-specific primers. microRNA was isolated from the cells or tissue using mirVana miRNA Isolation Kit following manufacturer’s protocol, and microRNA-specific cDNA synthesis was performed using TaqMan MicroRNA Reverse Transcription Kit. TaqMan Multiplex Master Mix was used to perform real-time quantitative PCR for miRNA with specific miRNA primers of miR-210-3p, miR-200b-3p, miR-126, and U6 sn RNA. mRNA and miRNA expression were normalized to β-actin and U6 snRNA, respectively, following the ΔΔCT method. Mean ΔCt value was transformed to relative expression or fold change by 2ΔΔCT and the average fold change value was calculated.
Immunoblotting
Total protein isolation from in vitro studies was performed following studies performed earlier by the group (46). Briefly, cells were lysed and centrifuged at 13,0000 rpm for 10 min at 4 °C. Protein concentrations of cell lysates were determined by the BCA Protein Assay Kit following manufactures’ guidelines. Protein isolation from the tissue was performed by weighing 300 mg of tissue and lyse in RIPA lysis buffer (for 3 times and 30 s each) and shaking in Tissue lyser (Qiagen). Cell lysates (50 μg of protein) were resolved on 10% SDS–PAGE and transferred onto PVDF membranes with the help of Turbo Blotting System (Bio-Rad Laboratories). Membranes were first blocked with 5% BSA in Tris-buffered saline buffer for 1 h followed by the overnight incubation with primary antibodies in a rotating shaker at 4 °C. The membranes were then washed three times with TBST (TBS containing 0.1% Tween 20) buffer for 10 min intervals and incubated with peroxidise-conjugated specific secondary antibodies for 2 h at room temperature. Membranes were then washed three times with TBST for 10 min intervals and subjected to ECL Substrate incubation for 5 min at room temperature. Protein bands were visualized in Chemidoc XRS+ System (Bio-Rad Laboratories, Hercules, California, USA) using Image Lab Software (https://imagej.nih.gov/ij/).
Enzyme-linked immunosorbent assay
We measured insulin levels in the cell culture medium of control and treated cells using mouse ELISA kits following the manufacturer’s instructions.
Flow cytometry
Visceral adipose tissues collected from human and mice were rinsed in sterile PBS, chopped into small pieces, and then digested in Hanks' Balanced Salt Solution containing collagenase type II (2 mg/ml), glucose (5.5 mM), and 4% BSA (fatty acid-free) for 45 min at 37 °C water bath shaker. The enzymatic activity was then neutralized by the addition of serum and the digestion mixture was passed through a cell strainer (pore size: 70 μm). The isolated cell suspension was subjected to centrifugation at 2000 rpm for 10 min. Cell pellet was washed twice with ice-cold PBS and the cells were sorted for F4/80+ (mouse) or CD64+ (human) antibody for the collection of ATMs to perform different experiments. The differentiation of bone marrow cells into BMDMs was confirmed by staining with F4/80 antibodies followed by acquiring in BD Accuri C6 Plus.
Reanalyses of publicly available snSeq dataset
Human white adipose tissue single cell dataset was used for the analysis (22). The RDS file obtained by the authors was loaded into R, processed using Seurat package (47). We considered the following criteria to filter the cells: firstly, cells originated from OAT tissue only, second being BMI as a defining criterion of obese condition (lean: BMI < 25 and obese: BMI > 30), and thirdly, selected only ASPC, adipocytes, and macrophage cells. Seurat was used to visualize the UMAP projection of the cells and split by obese condition. Additionally, we calculated the proportion of each of the three cell types (ASPC, adipocytes, macrophages) for lean and obese conditions and all the OAT samples. For the mouse dataset (22), objects were directly imported in R and handled using Seurat. Mouse dataset was filtered to contain only three cell types, ASPC, adipocyte, and macrophages We calculated the cell proportion of each cell type obtained from HFD and Chow (projected as ‘StndD’) diet-fed mouse.
Statistical analyses
Data represented as mean ± SD. Student’s t test, two-way ANOVA were used to determine statistical significance, and a p-value at the level of p <0.05 was considered significant. Student’s t test was used for the comparisons among two groups and two-way ANOVA analyses were considered for calculating significance when comparing multiple groups. All data analyses were performed using GraphPad Prism software (v.9.0; GraphPad Software, Inc.).
| Reagent or Resource | Source/Company | Cat. No. |
|---|---|---|
| Antibodies | ||
| GLUT4 | Cell Signaling Technology | #2213 |
| β-actin | Thermo Fisher Scientific | #AM4302 |
| Pref1 | Abcam | ab119930 |
| CD63 | Abclonal | A19023 |
| F4/80 | Abcam | ab6640 |
| FABP4 | Cell Signaling Technology | #50699 |
| Anti-Mouse IgG (Alexa Fluor 488 conjugated) | Cell Signaling Technology | #4408 |
| Anti-Rabbit IgG (Alexa Fluor 488 conjugated) | Thermo Fisher Scientific | #A-11034 |
| Anti-Rabbit IgG (Alexa Fluor 568 conjugated) | Thermo Fisher Scientific | #A-1101 |
| Anti-Goat IgG (Alexa Fluor 594 conjugated) | Thermo Fisher Scientific | #A-11058 |
| FITC anti-mouse F4/80 antibody | BioLegend | 123107 |
| PE anti-human CD68 Antibody | BioLegend | 333801 |
| TruStainFcX (anti-mouse CD16/32) | BioLegend | 156603 |
| TruStainFcX (anti-human CD16/32) | BioLegend | 422301 |
| HRP-conjugated Anti-Mouse IgG antibody | Sigma-Aldrich | #A9044 |
| HRP-conjugated Anti-Rabbit IgG antibody | Sigma-Aldrich | #A9169 |
| Chemicals, peptides, and recombinant proteins | ||
| Ampicillin | Sigma-Aldrich | A0166 |
| DMEM | Gibco | #11995073 |
| Insulin solution from bovine pancreas | Sigma-Aldrich | I0516 |
| Bovine calf serum | ATCC | ATCC30-2030 |
| Penicillin-Streptomycin solution | Gibco | #15140122 |
| FBS | Gibco | #10082147 |
| Dexamethasone | Sigma-Aldrich | D1756 |
| IBMX | Sigma-Aldrich | I5879 |
| Minimum Essential Medium | ATCC | 30-2003 |
| RPMI 1640 | Gibco | #A1049101 |
| mouse M-CSF1 recombinant protein | Thermo Fisher Scientific | #PMC2044 |
| DMEM | ATCC | 30-2002 |
| Oil Red O | Sigma-Aldrich | # O1391 |
| Mayer’s Hematoxylin | Thermo Fisher Scientific | # 72804 |
| Eosin Stain Solution | Himedia | #S007 |
| Lipofectamine RNAiMAX transfection reagent | Invitrogen | #13778030 |
| Invivofectamine 3.0 reagent | Invitrogen | IVF3005 |
| Lipofectamine LTX reagent with PLUS reagent | Invitrogen | #15338100 |
| NP40 Cell lysis buffer | Invitrogen | #FNN0021 |
| Halt Protease and Phosphatase Inhibitor Cocktail | Invitrogen | #78441 |
| Rodent Diet With 10 kcal% Fat and Carbohydrate | Research Diet | D12450K |
| Rodent Diet With 60 kcal% Fat | Research Diet | D12492 |
| TRIzol | Invitrogen | 15596018 |
| iScript cDNA Synthesis Kit | Bio-Rad | #1708891 |
| PowerUpTM SYBRTM Green Master Mix | Applied Biosystems | A25742 |
| ClarityTM Western ECL Substrate | Bio-Rad | #1705061 |
| TaqManTM Multiplex Master Mix | Applied Biosystems | #4461881 |
| PVDF membranes | Bio-Rad | 1620177 |
| collagenase type I | Sigma-Aldrich | SCR103 |
| Tissue Freezing Medium | Leica Biosystems | 14020108926 |
| Rapid Detection of Firefly Luciferase Activity | Promega | E1500 |
| VECTASHIELD Antifade Mounting Medium with DAPI | Vector Laboratories | H-1200 |
| Vybrant DiO Cell-Labeling Solution | Invitrogen | V22886 |
| T4 DNA Ligase | NEB | # M0202S |
| XbaI | NEB | #R0145S |
| Sall | NEB | #R3138S |
| Opti-MEM | GIBCO | # 31985-062 |
| Kreb’s Ringer Bicarbonate Buffer | HiMedia | #TL-1097 |
| RNase A | Thermo Scientific | EN0531 |
| ExoQuick-TC ULTRA | SBI | EXOTC50A-1 |
| ExoQuick ULTRA | SBI | EQULTRA-20A-1 |
| Total exosome RNA and protein isolation kit | Invitrogen | #4478545 |
| mirVana miRNA Isolation Kit | Invitrogen | AM1560 |
| TaqMan MicroRNA Reverse Transcription Kit | Applied Biosystems | 4366596 |
| BCA Protein Assay Kit | Thermo Fisher Scientific | #23225 |
| mouse insulin ELISA kits | Elabscience | E-EL-M1382 |
| QuickChange Lightning Multi Site-Directed Mutagenesis Kit | Agilent | #210515 |
| Glucose Uptake Cell-Based Assay Kit | Cayman | #600470 |
| GeneJET Gel Extraction Kit | Thermo Fisher Scientific | K0691 |
| Cell lines | ||
| 3T3-L1 | ATCC | ATCCL-173 |
| C2C12 | ATCC | CRL-1772 |
| RAW264.7 | National Centre for Cell Science, Pune | N/A |
| Mouse | ||
| C57BL/6 | NIPER SAS Nagar | N/A |
| Oligonucleotides | ||
| miRIDIAN miR-210-3p mimic | Dharmacon | cat. no. #C-310570-05-0005 accession no.: MIMAT0000658 |
| miRIDIAN miR-210-3p inhibitor | Dharmacon | cat. no. #IH-310570-07-0005, accession no.:MIMAT0000267 |
| miRIDIAN miRNA hairpin mimic negative control | Dharmacon | cat. no. #CN-001000-01-05 accession no.:MIMAT0000039 |
| miRIDIAN miRNA hairpin inhibitor negative control | Dharmacon | cat. no. #IN-001005-01-05, accession no.: MIMAT0000039 |
| miRCURY miR-210-3p mimic LNA (5′FAM) | Qiagen | GeneGlobe ID: YM00470861-AGB; cat.no. 339174 |
| miRCURY control mimic LNA | Qiagen | GeneGlobe ID: YM00479902-AGA; cat. no. 339174 |
| miRCURY miR-210-3p mimic LNA | Qiagen | GeneGlobe ID: YM00470861-AGA, cat. no. 339174 |
| miRCURY mmu-miR-210-3p Inhibitor LNA (3′FAM) | Qiagen | GeneGlobe ID: YI04103147-DDC; catalog no. 339131 |
| miRCURY LNA miRNA Inhibitor Control | Qiagen | GeneGlobe ID: YI00199006-DDA; cat no. 339136 |
| Primers for qRT-PCR: See Table S1 | ||
| Recombinant DNA | ||
| pGL3-MYB-3′UTR | Addgene | #25798 RRID: Addgene_25798 |
| Software | ||
| ImageJ | Schneider et al. (48) | https://imagej.nih.gov/ij/ |
| GraphPad Prism 9 | GraphPad Software | https://www.graphpad.com/ |
| RNAhybrid | Rehmsmeier et al. (49) | https://bibiserv.cebitec.uni-bielefeld.de/rnahybrid;jsessionid=57c94cffe9f53afa1dc320ba3c06 |
| NTA 3.3 | Malvern Panalytical | https://www.malvernpanalytical.com/en/support/product-support/software/nanosight-nta-software-update-v3-3 |
| LasX | Leica Microsystem | https://www.leica-microsystems.com/products/microscope-software/p/leica-las-x-ls/?nlc=20221205-SFDC-015771&utm_source=google&utm_medium=cpc&utm_campaign=23-AP-ALL-L3-GLMS-GOOG-PP-India-General-Search&utm_content=text_ad&utm_term=leica%20lasx&gad_source=1&gclid=CjwKCAiA98WrBhAYEiwA2WvhOl1xYTWBcJdjT2GCRz-AEOFnU0_-_Z_s554tEkPXdZyioJxb-IBRkRoC_uYQAvD_BwE |
| Zen Black | Carl Zeiss | https://www.micro-shop.zeiss.com/en/de/softwarefinder/software-categories/zen-black/zen-black-system/ |
Data availability
D.P. is the guarantor of this work and, as such, has full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We express our gratitude to the NIPER SAS Nagar, Animal House Facility for mice experiments and to the IIT Ropar for providing instrumentation facilities. We are also thankful to Dr Neelkanth Nirmalkar, Department of Chemical Engineering, IIT Ropar for his kind help in Nanosight LM10 NTA-based analysis. This study was supported by the SERB-Early Career Research Grant (Project No.: ECR/2017/000892), Govt. of India and the DBT-Twining project (Project No.:BT/PR24700/NER/95/819/2017), Govt. of India Debarun Patra and P. R. acknowledges the IIT Ropar and the Ministry of Education, Govt. of India for their research fellowships.
Author contributions
Debarun Patra and Durba Pal conceptualization; Debarun Patra, P. R., Shivam Sharma, U. D., V. K., and Satpal Singh data curation; Debarun Patra, S. D., and Durba Pal formal analysis; Debarun Patra, P. R., Shivam Sharma, U. D., V. K., and Satpal Singh investigation; Debarun Patra and Durba Pal methodology; Debarun Patra, P. R., Shivam Sharma, V. K., and Satpal Singh validation; Debarun Patra and P. R. visualization; Debarun Patra and Durba Pal writing–original draft; Debarun Patra, S. D., K. T., and Durba Pal writing–review and editing; U. D. and A. K. software; Satpal Singh, S. D., K. T., A. K., Durba Pal resources; Durba Pal supervision; Durba Pal funding acquisition.
Reviewed by members of the JBC Editorial Board. Edited by Qi-Qun Tang
Supporting information
References
- 1.Hruby A., Hu F.B. The epidemiology of obesity: a big picture. Pharmacoeconomics. 2015;33:673–689. doi: 10.1007/s40273-014-0243-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fernández-Real J.M., Ricart W. Insulin resistance and chronic cardiovascular inflammatory syndrome. Endocr. Rev. 2003;24:278–301. doi: 10.1210/er.2002-0010. [DOI] [PubMed] [Google Scholar]
- 3.Grant R.W., Dixit V.D. Adipose tissue as an immunological organ: adipose tissue as an immunological organ. Obesity. 2015;23:512–518. doi: 10.1002/oby.21003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reilly S.M., Saltiel A.R. Adapting to obesity with adipose tissue inflammation. Nat. Rev. Endocrinol. 2017;13:633–643. doi: 10.1038/nrendo.2017.90. [DOI] [PubMed] [Google Scholar]
- 5.Ye J. Emerging role of adipose tissue hypoxia in obesity and insulin resistance. Int. J. Obes. 2009;33:54–66. doi: 10.1038/ijo.2008.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goossens G.H., Bizzarri A., Venteclef N., Essers Y., Cleutjens J.P., Konings E., et al. Increased adipose tissue oxygen tension in obese compared with lean men is accompanied by insulin resistance, impaired adipose tissue capillarization, and inflammation. Circulation. 2011;124:67–76. doi: 10.1161/CIRCULATIONAHA.111.027813. [DOI] [PubMed] [Google Scholar]
- 7.McNelis J.C., Olefsky J.M. Macrophages, immunity, and metabolic disease. Immunity. 2014;41:36–48. doi: 10.1016/j.immuni.2014.05.010. [DOI] [PubMed] [Google Scholar]
- 8.Pal D., Dasgupta S., Kundu R., Maitra S., Das G., Mukhopadhyay S., et al. Fetuin-A acts as an endogenous ligand of TLR4 to promote lipid-induced insulin resistance. Nat. Med. 2012;18:1279–1285. doi: 10.1038/nm.2851. [DOI] [PubMed] [Google Scholar]
- 9.Maniyadath B., Zhang Q., Gupta R.K., Mandrup S. Adipose tissue at single-cell resolution. Cell Metab. 2023;35:386–413. doi: 10.1016/j.cmet.2023.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jaitin D.A., Adlung L., Thaiss C.A., Weiner A., Li B., Descamps H., et al. Lipid-associated macrophages control metabolic homeostasis in a trem2-dependent manner. Cell. 2019;178:686–698.e14. doi: 10.1016/j.cell.2019.05.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hill D.A., Lim H.-W., Kim Y.H., Ho W.Y., Foong Y.H., Nelson V.L., et al. Distinct macrophage populations direct inflammatory versus physiological changes in adipose tissue. Proc. Natl. Acad. Sci. U. S. A. 2018 doi: 10.1073/pnas.1802611115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chakarov S., Blériot C., Ginhoux F. Role of adipose tissue macrophages in obesity-related disorders. J. Exp. Med. 2022;219 doi: 10.1084/jem.20211948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeppesen D.K., Zhang Q., Franklin J.L., Coffey R.J. Extracellular vesicles and nanoparticles: emerging complexities. Trends Cell Biol. 2023;33:667–681. doi: 10.1016/j.tcb.2023.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Théry C., Zitvogel L., Amigorena S. Exosomes: composition, biogenesis and function. Nat. Rev. Immunol. 2002;2:569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 15.Crewe C., Funcke J.-B., Li S., Joffin N., Gliniak C.M., Ghaben A.L., et al. Extracellular vesicle-based interorgan transport of mitochondria from energetically stressed adipocytes. Cell Metab. 2021;33:1853–1868.e11. doi: 10.1016/j.cmet.2021.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crewe C., Joffin N., Rutkowski J.M., Kim M., Zhang F., Towler D.A., et al. An endothelial-to-adipocyte extracellular vesicle axis governed by metabolic state. Cell. 2018;175:695–708.e13. doi: 10.1016/j.cell.2018.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pan Y., Hui X., Hoo R.L.C., Ye D., Chan C.Y.C., Feng T., et al. Adipocyte-secreted exosomal microRNA-34a inhibits M2 macrophage polarization to promote obesity-induced adipose inflammation. J. Clin. Invest. 2019;129:834–849. doi: 10.1172/JCI123069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ying W., Riopel M., Bandyopadhyay G., Dong Y., Birmingham A., Seo J.B., et al. Adipose tissue macrophage-derived exosomal miRNAs can modulate in vivo and in vitro insulin sensitivity. Cell. 2017;171:372–384.e12. doi: 10.1016/j.cell.2017.08.035. [DOI] [PubMed] [Google Scholar]
- 19.Tang Y., Yang L.-J., Liu H., Song Y.-J., Yang Q.-Q., Liu Y., et al. Exosomal miR-27b-3p secreted by visceral adipocytes contributes to endothelial inflammation and atherogenesis. Cell Rep. 2023;42 doi: 10.1016/j.celrep.2022.111948. [DOI] [PubMed] [Google Scholar]
- 20.Amano S.U., Cohen J.L., Vangala P., Tencerova M., Nicoloro S.M., Yawe J.C., et al. Local proliferation of macrophages contributes to obesity-associated adipose tissue inflammation. Cell Metab. 2014;19:162–171. doi: 10.1016/j.cmet.2013.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boutens L., Stienstra R. Adipose tissue macrophages: going off track during obesity. Diabetologia. 2016;59:879–894. doi: 10.1007/s00125-016-3904-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Emont M.P., Jacobs C., Essene A.L., Pant D., Tenen D., Colleluori G., et al. A single-cell atlas of human and mouse white adipose tissue. Nature. 2022;603:926–933. doi: 10.1038/s41586-022-04518-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patra D., Roy S., Arora L., Kabeer S.W., Singh S., Dey U., et al. miR-210-3p promotes obesity-induced adipose tissue inflammation and insulin resistance by targeting SOCS1-mediated NF-κB pathway. Diabetes. 2023;72:375–388. doi: 10.2337/db22-0284. [DOI] [PubMed] [Google Scholar]
- 24.Salomon C., Das S., Erdbrügger U., Kalluri R., Kiang Lim S., Olefsky J.M., et al. Extracellular vesicles and their emerging roles as cellular messengers in endocrinology: an endocrine society scientific statement. Endocr. Rev. 2022;43:441–468. doi: 10.1210/endrev/bnac009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taylor D.D., Zacharias W., Gercel-Taylor C. In: Serum/Plasma Proteomics: Methods and Protocols. Simpson R.J., Greening D.W., editors. Humana Press; Totowa, NJ: 2011. Exosome isolation for proteomic analyses and RNA profiling; pp. 235–246. [DOI] [PubMed] [Google Scholar]
- 26.Wang Y., Zhao M., Liu S., Guo J., Lu Y., Cheng J., et al. Macrophage-derived extracellular vesicles: diverse mediators of pathology and therapeutics in multiple diseases. Cell Death Dis. 2020;11:1–18. doi: 10.1038/s41419-020-03127-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Akbar N., Azzimato V., Choudhury R.P., Aouadi M. Extracellular vesicles in metabolic disease. Diabetologia. 2019;62:2179–2187. doi: 10.1007/s00125-019-05014-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arcidiacono B., Chiefari E., Foryst-Ludwig A., Currò G., Navarra G., Brunetti F.S., et al. Obesity-related hypoxia via miR-128 decreases insulin-receptor expression in human and mouse adipose tissue promoting systemic insulin resistance. EBioMedicine. 2020;59 doi: 10.1016/j.ebiom.2020.102912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu G., Ji C., Song G., Zhao C., Shi C., Song L., et al. MiR-26b modulates insulin sensitivity in adipocytes by interrupting the PTEN/PI3K/AKT pathway. Int. J. Obes. 2015;39:1523–1530. doi: 10.1038/ijo.2015.95. [DOI] [PubMed] [Google Scholar]
- 30.Trajkovski M., Hausser J., Soutschek J., Bhat B., Akin A., Zavolan M., et al. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature. 2011;474:649–653. doi: 10.1038/nature10112. [DOI] [PubMed] [Google Scholar]
- 31.Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.György B., Hung M.E., Breakefield X.O., Leonard J.N. Therapeutic applications of extracellular vesicles: clinical promise and open questions. Annu. Rev. Pharmacol. Toxicol. 2015;55:439–464. doi: 10.1146/annurev-pharmtox-010814-124630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ismail N., Wang Y., Dakhlallah D., Moldovan L., Agarwal K., Batte K., et al. Macrophage microvesicles induce macrophage differentiation and miR-223 transfer. Blood. 2013;121:984–995. doi: 10.1182/blood-2011-08-374793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raposo G., Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J. Cell Biol. 2013;200:373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bobrie A., Colombo M., Raposo G., Théry C. Exosome secretion: molecular mechanisms and roles in immune responses. Traffic. 2011;12:1659–1668. doi: 10.1111/j.1600-0854.2011.01225.x. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Y., Shi L., Mei H., Zhang J., Zhu Y., Han X., et al. Inflamed macrophage microvesicles induce insulin resistance in human adipocytes. Nutr. Metab. (Lond) 2015;12:21. doi: 10.1186/s12986-015-0016-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ying W., Gao H., Dos Reis F.C.G., Bandyopadhyay G., Ofrecio J.M., Luo Z., et al. MiR-690, an exosomal-derived miRNA from M2-polarized macrophages, improves insulin sensitivity in obese mice. Cell Metab. 2021;33:781–790.e5. doi: 10.1016/j.cmet.2020.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ortega F.J., Moreno M., Mercader J.M., Moreno-Navarrete J.M., Fuentes-Batllevell N., Sabater M., et al. Inflammation triggers specific microRNA profiles in human adipocytes and macrophages and in their supernatants. Clin. Epigenetics. 2015;7:49. doi: 10.1186/s13148-015-0083-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thomou T., Mori M.A., Dreyfuss J.M., Konishi M., Sakaguchi M., Wolfrum C., et al. Adipose-derived circulating miRNAs regulate gene expression in other tissues. Nature. 2017;542:450–455. doi: 10.1038/nature21365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saltiel A.R. Insulin signaling in health and disease. J. Clin. Invest. 2021;131 doi: 10.1172/JCI142241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parks B.W., Sallam T., Mehrabian M., Psychogios N., Hui S.T., Norheim F., et al. Genetic architecture of insulin resistance in the mouse. Cell Metab. 2015;21:334–347. doi: 10.1016/j.cmet.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pant R., Kabeer S.W., Sharma S., Kumar V., Patra D., Pal D., et al. Pharmacological inhibition of DNMT1 restores macrophage autophagy and M2 polarization in Western diet–induced nonalcoholic fatty liver disease. J. Biol. Chem. 2023;299 doi: 10.1016/j.jbc.2023.104779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Navarro F., Gutman D., Meire E., Cáceres M., Rigoutsos I., Bentwich Z., et al. miR-34a contributes to megakaryocytic differentiation of K562 cells independently of p53. Blood. 2009;114:2181–2192. doi: 10.1182/blood-2009-02-205062. [DOI] [PubMed] [Google Scholar]
- 44.Banerjee D., Patra D., Sinha A., Roy S., Pant R., Sarmah R., et al. Lipid-induced monokine cyclophilin-A promotes adipose tissue dysfunction implementing insulin resistance and type 2 diabetes in zebrafish and mice models of obesity. Cell. Mol. Life Sci. 2022;79:282. doi: 10.1007/s00018-022-04306-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arora L., Patra D., Roy S., Nanda S., Singh N., Verma A.K., et al. Hypoxia-induced miR-210-3p expression in lung adenocarcinoma potentiates tumor development by regulating CCL2-mediated monocyte infiltration. Mol. Oncol. 2024;18:1278–1300. doi: 10.1002/1878-0261.13260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Choudhary S.A., Patra D., Sinha A., Mazumder S., Pant R., Chouhan R., et al. A small molecule potent IRAK4 inhibitor abrogates lipopolysaccharide-induced macrophage inflammation in-vitro and in-vivo. Eur. J. Pharmacol. 2023;944 doi: 10.1016/j.ejphar.2023.175593. [DOI] [PubMed] [Google Scholar]
- 47.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rehmsmeier M., Steffen P., Höchsmann M., Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
D.P. is the guarantor of this work and, as such, has full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.