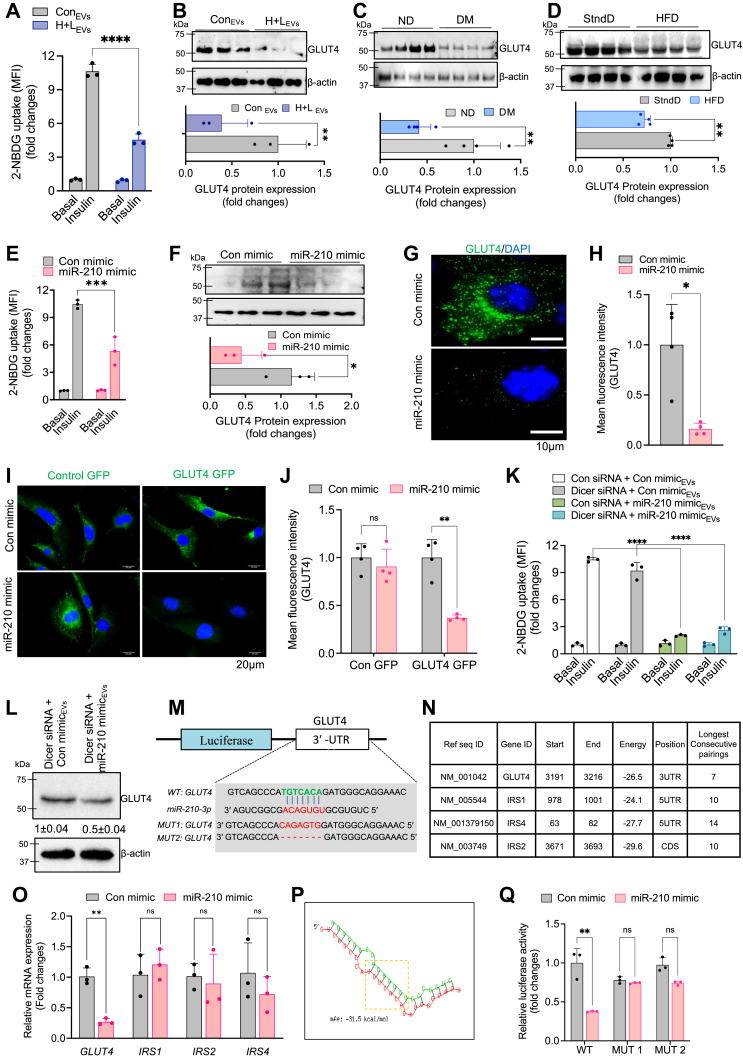
Figure 2.
Loss of glycemic homeostasis associated with direct suppression of GLUT4 mediated by obese ATMs-driven miR-210-3p.A, insulin-stimulated glucose uptake was monitored by measuring 2-NBDG in the cell lysate of adipocytes, incubated with control, and H + L–treated BMDM released EVs. (n = 3); ∗∗∗∗p < 0.0001. B–D, Western blot analysis and quantification of GLUT4 protein expression in adipocytes exposed with control or H + L–incubated BMDM EVs (B); adipocytes isolated from VAT of ND and DM patients (C); adipocytes isolated from VAT of SD and HFD mice (D). E, glucose uptake in the presence or absence of insulin stimulation in control mimic or miR-210-3p mimic transfected adipocytes (n = 3); ∗∗∗p < 0.001. F, Western blot analysis showed GLUT4 expression in adipocytes transfected with control mimic and miR-210-3p mimic (n = 3), (n = 3); ∗∗p < 0.01 by Student’s t test. G and H, GLUT4 immunofluorescence imaging (G) and quantification analyses (H) in adipocytes transfected with control mimic and miR-210-3p mimic (n = 3). Scale bar represents 10 μm. ∗∗p < 0.01 by Student’s t test. I and J, immunofluorescence image showing GLUT4 migration in murine myoblast cells transfected with control mimic or miR-210-3p mimic (I) and quantification analysis (J) (n = 3); ∗∗p < 0.01, ns= not significant, by two-way ANOVA. K and L, glucose uptake assay (K) and western blotting analyses of GLUT4 (L) in adipocytes under the presence or absence of insulin stimulation in adipocytes cocultured with Dicer-silenced macrophage transfected with control mimic or miR-210-3p mimic. Quantification analyses, (n = 3), ∗∗∗∗p < 0.0001 by student’s t test. M, miRNA target prediction analyses using miRWALK database search analysis showed miR-210-3p targets for insulin signaling pathways molecule. N, mRNA expression analyses of predicted targets GLUT4, IRS1, IRS2, IRS4 in adipocytes transfected with control mimic or miR-210-3p mimic, (n = 3), ∗∗p < 0.01, ns-not significant, by student’s t test. O, the putative-binding site of GLUT4 with miR-210-3p and seed sequence of miR-210-3p and mutated 3′-UTR site of GLUT4. P, RNAhybrid webserver was used to predict the minimum free energy (mfe) of GLUT4 mRNA transcripts and miRNA-210-3p interactions. Q, luciferase activity in adipocytes transfected with WT or mutated (mut1 and mut 2) GLUT4 plasmid constructs and control mimic or miR-210-3p mimic (n = 3). ∗∗p < 0.01 by Student’s t test. Data are expressed as means ± SD; n = 3–5; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ns = non-significant (Student’s t test and two-way ANOVA). 2-NBDG, 2-(N-(7-Nitrobenz-2-oxa-1,3-diazol-4-yl)Amino)-2-Deoxyglucose; ATM, adipose tissue macrophage; BMDM, bone marrow-dervied macrophage; Con mimic, control mimic; EVs, extracellular vesicles; EV, extracellular vesicle; H, hypoxia; L, lipid; ND, non-obese non-diabetic; ns, not significant.