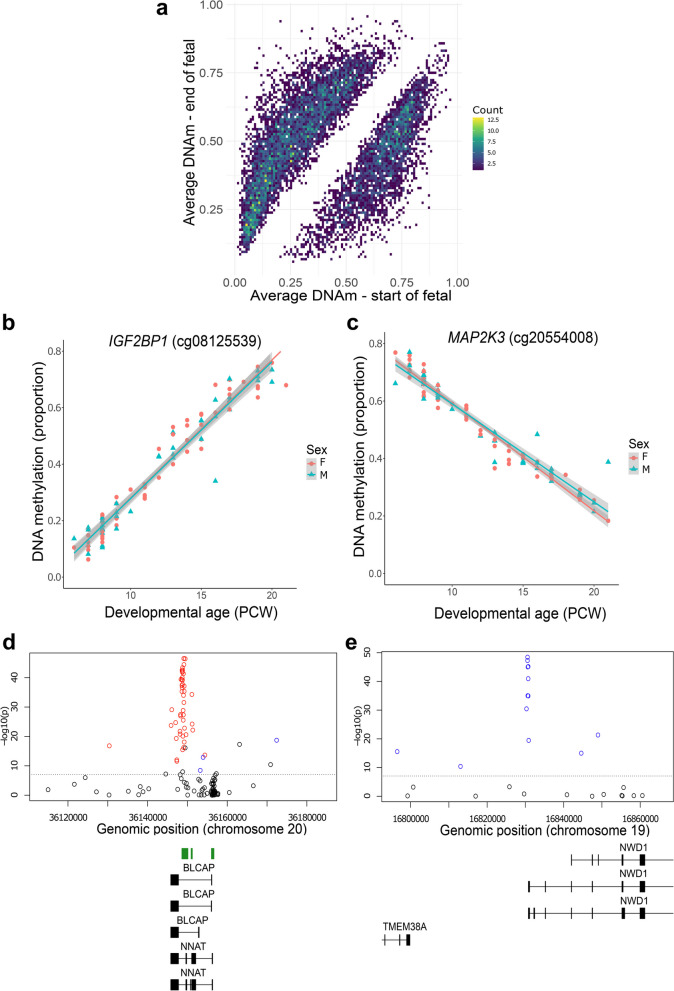
Fig. 2.
Changes in DNA methylation associated with development of the human pancreas. A Mean levels of DNA methylation in early fetal pancreas samples (6—8 PCW) and later fetal pancreas samples (19—21 PCW) across all significant dDMPs. B cg08125539, annotated to the IGF2BP1 gene on chromosome 17, was the most significant hypermethylated dDMP across pancreas development (change in DNA methylation proportion per gestational week = 0.049, P = 1.35 × 10–61). C cg20554008, annotated to the MAP2K3 gene on chromosome 17, was the most significant hypomethylated dDMP across pancreas development (change in DNA methylation proportion per gestational week = -0.036, P = 1.48 × 10–61). Many dDMPs are colocalized into larger regions of differential DNA methylation (dDMRs) associated with pancreas development. Shown are (D) the top-ranked hypermethylated dDMR located in the BLCAP and NNAT genes on chromosome 20 (regression coefficient = 0.253, P = < 1.14 × 10–304) and E) the top-ranked hypomethylated dDMR located in the transcription start site of NWD1 on chromosome 19 (regression coefficient = -0.239, P = P = < 1.14 × 10–304)