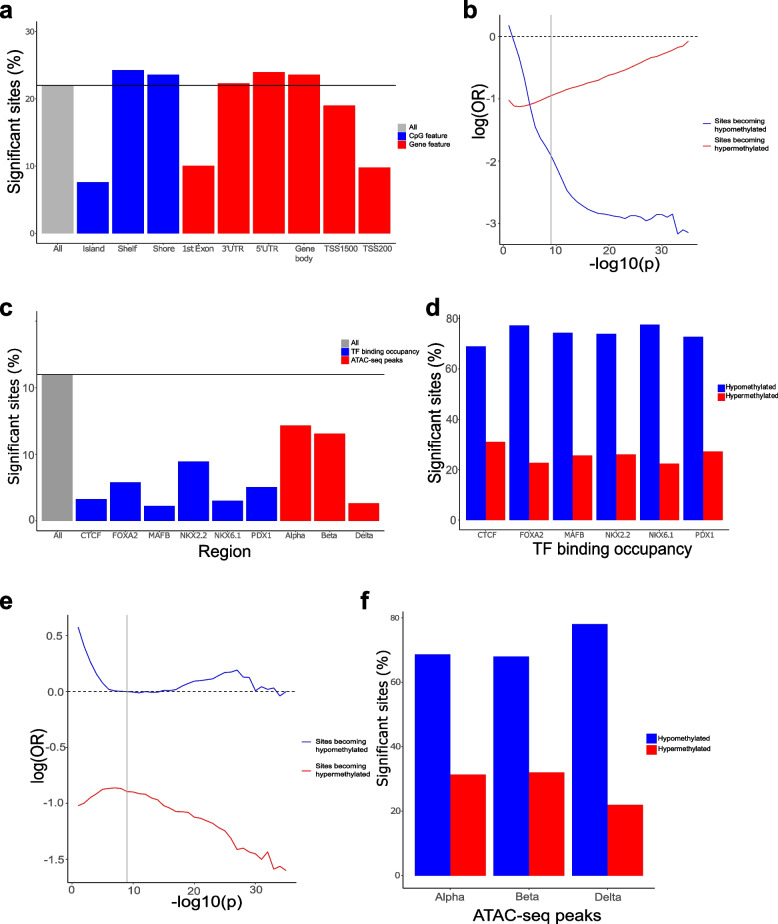
Fig. 4.
The distribution and direction of fetal pancreas dDMPs differs markedly across genomic features. A Compared to the genome average, dDMPs are significantly underrepresented in CpG islands, transcription start-sites, 5′ UTRs, and first exons, but significantly enriched in CpG island shores, CpG island shelves, and the gene body. Relative enrichment of dDMPs in CpG island features. B The depletion of dDMPs in CpG islands is largely driven by a dramatic paucity of sites becoming hypomethylated across development. Sites becoming hypomethylated across development are shown in blue and sites becoming hypermethylated across development are shown in red. Shown is the relative enrichment of sites in each category based on their P-value for association with developmental age (from 1 × 10–1, to 1 × 10–35) determined using a Fisher’s exact test. The genome-wide significance threshold for dDMPs (P < 9 × 10–8) is denoted using a grey line. C Compared to the genome average, dDMPs are also depleted in regulatory domains defined by key transcription factor binding site occupancy and chromatin accessibility in pancreatic islets from human donors. In contrast to the pattern seen in CpG islands, these regulatory domains are characterized by a dramatic depletion of hypermethylated dDMPs for both regions of (D) transcription factor binding occupancy and (E) open chromatin in pancreatic islet cell-types. F The depletion of dDMPs in domains bound by CTCF, for example, is largely driven by a dramatic paucity of sites becoming hypomethylated across development