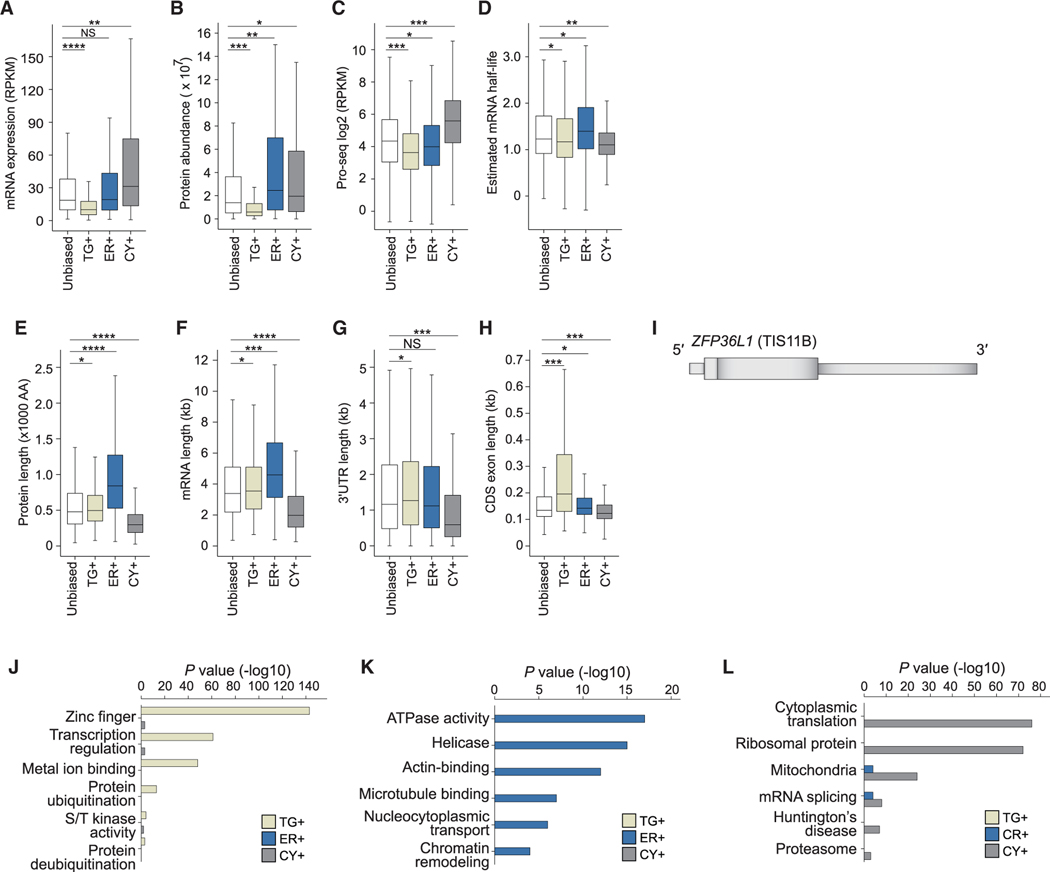
Figure 2. Characteristics of compartment-enriched mRNAs.
(A) Steady-state mRNA abundance levels obtained from whole-cell lysates. TG+, N = 1,246; ER+, N = 919, CY+, N = 1,481; unbiased, N = 3,369. Mann-Whitney test: *0.05 > p > 10−9; **10−10 >p> 10−20; ***10−21 >p> 10−80; ****10−81 > p > 0. Exact p values are listed in Table S3. Boxplots depict median, 25th and 75th percentiles (box), and 5% and 95% confidence intervals (error bars).
(B) As in (A), but steady-state protein levels obtained from whole-cell lysates are shown. TG+, N = 469; ER+, N = 638; CY+, N = 833; unbiased, N = 2,001.
(C) As in (B), but Pro-seq levels are shown, which indicate transcription rates. TG+, N = 1,222; ER+, N = 896; CY+, N = 1,425; unbiased, N = 3,268.
(D) As in (C), but estimated mRNA half-lives are shown.
(E) As in (A), but protein size distributions are shown. AA, amino acid.
(F) As in (A), but mRNA length distributions are shown.
(G) As in (A), but 3′ UTR length distributions are shown.
(H) As in (A), but average CDS exon length distributions are shown.
(I) ZFP36L1 (TIS11B) mRNA model. Tall boxes: CDS exons, narrow boxes: 5′ and 3′ UTRs.
(J) Gene ontology analysis for TG+ mRNAs. Top six functional gene classes uniquely enriched in TG+ mRNAs and Benjamini Hochberg-adjusted p values are shown.
(K) As in (J), but for ER+ mRNAs.
(L) As in (J), but for CY+ mRNAs.