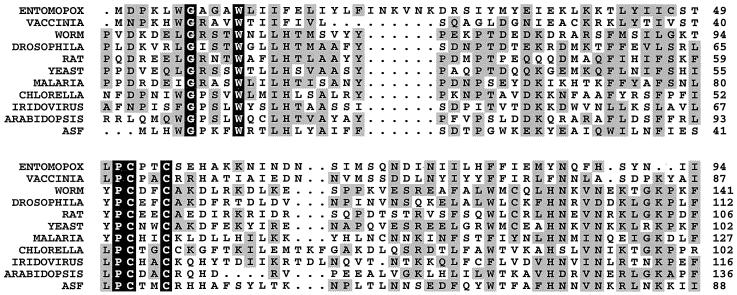
FIG. 1.
Alignment of the predicted amino acid sequence of the ASFV Malawi Lil-20/1 ORF 9GL product (ASF) with the yeast ERV1 and rat ALR motif-containing proteins. Homologues shown are as follows (with the GenBank accession number of the nucleotide sequence given in parentheses following each): ENTOMOPOX, M. sanguinipes entomopox virus ORF MSV093 product (AF063866); VACCINIA, vaccinia virus ORF E10R product (M35027); WORM, C. elegans gene product F56C11.3 (AF043697); DROSOPHILA, D. melanogaster homologue (AA696037); RAT, Rattus norvegicus ALR (D30735); YEAST, S. cerevisiae ERV1 (X60722); MALARIA, P. falciparum homologue (AL031745); CHLORELLA, chlorella virus ORF A465R product (U42580); IRIDOVIRUS, fish lymphocystis disease virus homologue (L63545); ARABIDOPSIS, Arabidopsis thaliana (AC003058). The consensus sequence was constructed with the Clustal W computer program (58) by using the Dayhoff PAM 250 symbol comparison table with a 0.5 cutoff value. Invariant amino acids in all proteins are shown on a solid background, while conservative substitutions are shaded. Periods in the sequence denote gaps introduced by the alignment program.