Figure 1.
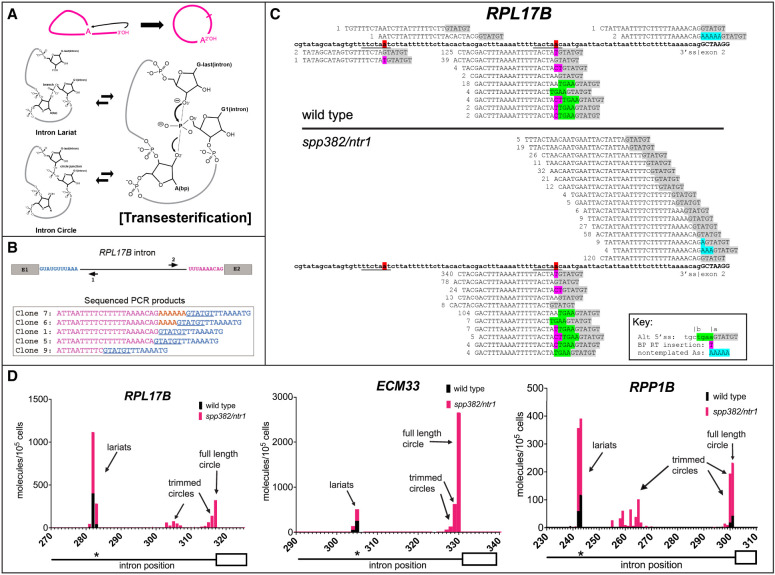
A hypothesis and detection of intron circles in S. cerevisiae. (A) A possible mechanism for the spliceosome to create intron circles by nucleophilic attack of the lariat tail 3′-OH. (Top) Conversion of the lariat to a circle, releasing the 2′-OH of the A residue from the branch (arrow). (Bottom) Proposed chemical mechanism for attack of the lariat tail on the phosphate at the branch (bold P), starting with the lariat (top left) and then the transition state for the transesterification reaction (right), followed by circle formation (bottom left). The reactions are reversible. (B) RPL17B intron with “inverse” primers to capture circular intron junctions. Sequences of selected cloned PCR products are shown, with the 5′ss underlined and nongenomic As shown in gold. (C) High-throughput sequencing reveals a complex set of processed circular RPL17B introns whose abundance is dramatically increased by the spp382-1 mutation. The intron is in bold. Reads are aligned above (circles) or below (lariats), the 5′ss sequence is highlighted in gray, and nongenomic As are in blue. Deduplicated (unique) read counts from each library are shown in front of each read. Lariats derived from incorrect 5′ splice sites are in green. (D) Different introns produce distinct distributions of intron circles. Graphs show the junction locations (x-axis) and number of unique reads with junctions at that location (y-axis; calibrated to a spiked-in circular RNA) per 105 yeast cells from wild-type (black bars) or spp382-1 (pink bars) cells. The 3′ part of each intron (line) and its second exon (white box) are shown below the x-axis, with the asterisk indicating the position of the BP. Additional introns and data are shown in Supplemental Figure S1 and Supplemental Table S1.