Figure 2.
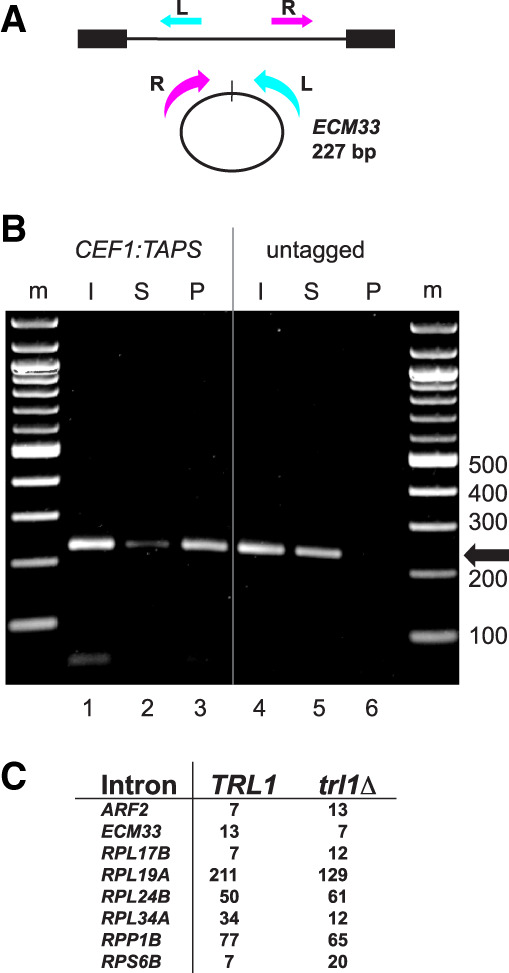
Intron circles are associated with the spliceosome and are not formed by the tRNA ligase Trl1. (A) Detecting circular ECM33 introns. Inverse PCR primers that create a 227 bp product from full-length ECM33 intron circles are shown; the R primer is 3′ of the branchpoint and cannot detect lariats. (B) Association of ECM33 intron circles with the spliceosome in spp382-1 cells. The spp382-1 mutant was fitted with a TAPS tag at the C terminus of its CEF1-coding region for affinity purification of spliceosomes. Extracts from this strain and an untagged control were prepared (input [I]) and bound in batches to IgG-sepharose. Unbound (supernatant [S]) and bound (pellet [P]) fractions were prepared. RNA from each fraction was used for RT-PCR to identify ECM33 circles (227 bp product). The pellet fraction from the tagged (but not the untagged) extract was enriched by primer extension in spliceosomal U2 snRNA and free of cytoplasmic SCR1 (Supplemental Fig. S2). (C) Robust detection of intron circles in a strain lacking tRNA ligase. The table shows spike-in-normalized circular intron counts for eight introns from libraries prepared from strains that differ by deletion of TRL1. Detailed data are shown in Supplemental Table S2.
