Figure 3.
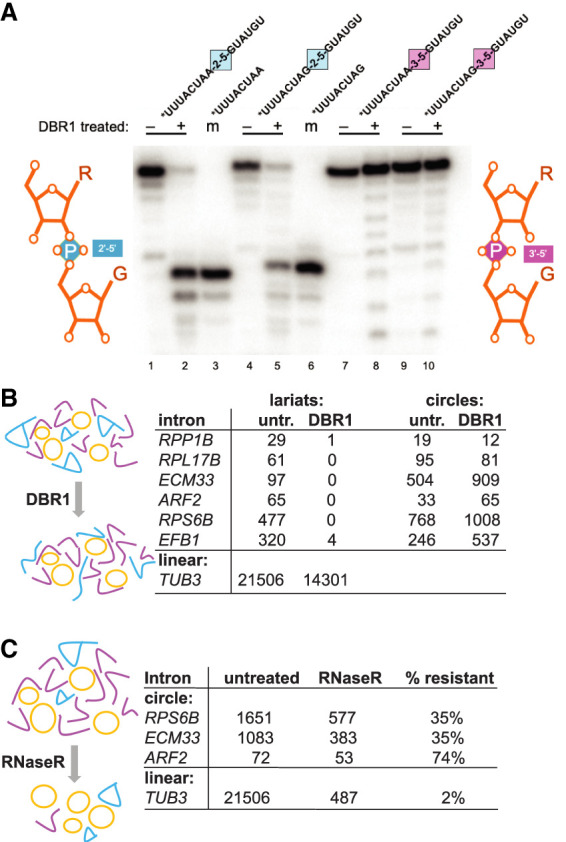
Enzymatic tests of circular intron structure. (A) Dbr1 does not require a 3′ nucleotide at the branch and can cleave an internal 2′–5′ linkage. 32P-5′ end-labeled RNA oligonucleotides carrying either a single 2′–5′ internal linkage in the middle of an otherwise 3′–5′-linked RNA or carrying only 3′–5′-linked nucleotides were incubated (or not) with recombinant Dbr1 and separated by electrophoresis on an 8 M urea/20% acrylamide gel. Two different substrate sequences (bpA-G5ss and 3ssG-G5ss) were made with either a 2′–5′ (blue) or a 3′–5′ (pink) linkage at the junction (marked by the colored squares) and are shown above each reaction. Minus and plus signs indicate enzyme addition. (Lanes 3,6) Two 8-mers identical to the expected Dbr1 cleavage products are labeled as markers (m). Comigration with the marker indicates cleavage at the 2′–5′ linkage. (B) Circle junctions from the spp382-1 mutant are resistant to cleavage with Dbr1, whereas lariat junctions are not. Read counts of lariats and circles for six introns from libraries made with or without prior treatment of the input RNA with Dbr1 are shown. (C) Circle junction-containing RNAs from the spp382-1 mutant are much more resistant to RNase R than a linear pre-mRNA. Read counts of circles for three introns from libraries made with or without prior treatment of the input RNA with RNase R are shown. Detailed data are shown in Supplemental Table S3.
