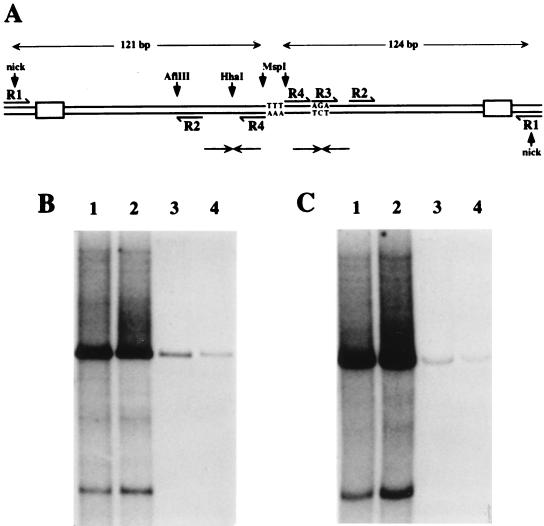
FIG. 2.
(A) Schematic representation of the tetramer bridge region of RF DNA, created by unfolding and copying the right-end hairpin sequence shown in Fig. 1A. The unpaired nucleotides at the top of the stem (TTT) now form the dyad symmetry axis of a palindromic duplex junction. Pairs of small opposing arrows indicate the positions of the internal palindromes detailed in Fig. 1C. Sequences used as primers for PCR-mediated subcloning of this region are labeled R1-R4. Shaded boxes near the nick sites represent the inboard NS1 binding sites. (B and C) Autoradiographs of native agarose gels showing ScaI-linearized products of NS1-driven in vitro replication assays programmed with plasmids containing inserts derived from single arms of the bridge before (B) and after (C) immunoprecipitation with anti-NS1 serum. Replication substrates were pR1-4AGA (lane 1), pR1-4 Hha (lane 2), pR1-2 (lane 3), and pR1-3 (lane 4). Autoradiographs were overexposed to show residual levels of replication seen with defective substrates. In all cases, digestion with ScaI reduced replication products to a single major species which comigrated with unit-length plasmid substrate DNA. In addition, such long exposures revealed a minor, smaller product and multiple rarer species, none of whose structures are currently understood.