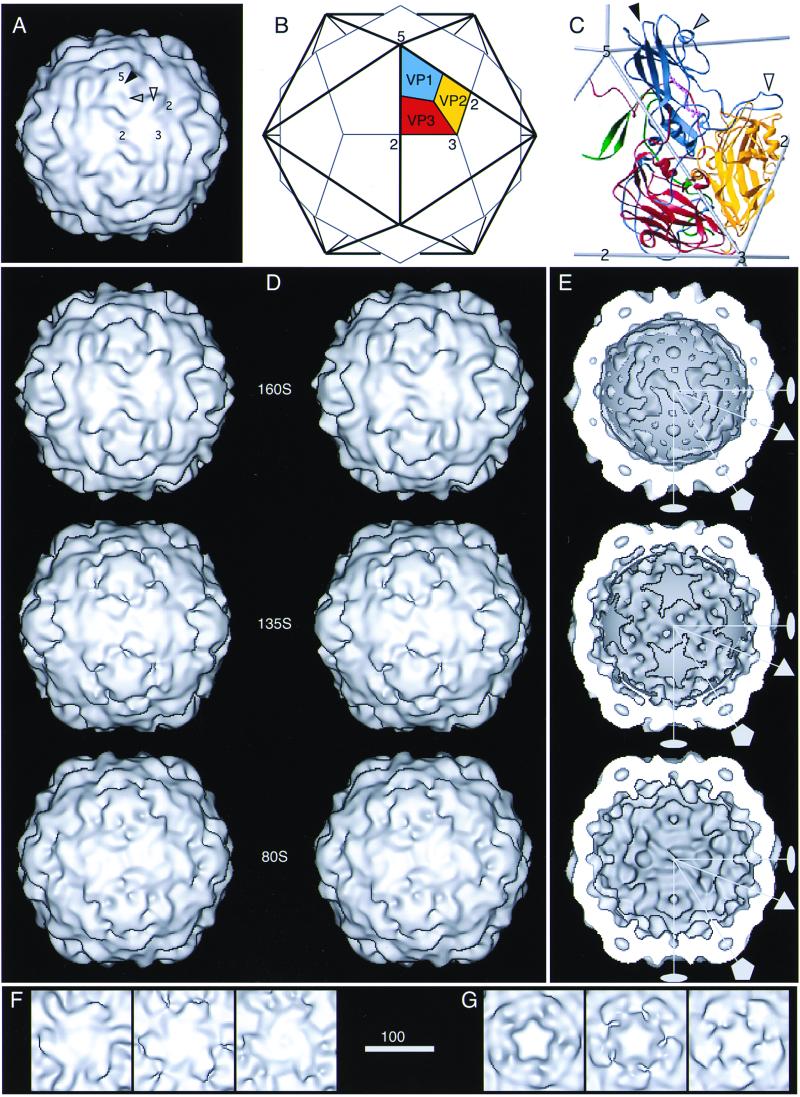
FIG. 2.
(A to C) Depiction of the 160S poliovirus particle as visualized by X-ray crystallography (31). One fivefold, one threefold, and two twofold symmetry axes are labeled. (A) Surface rendering of the outer surface, after the structure was limited to 22 Å by the procedure described (7). In panels B and C, the location of one protomer—consisting of one copy of VP1 (blue), VP2 (yellow), VP3 (red), and VP4 (green; shown only in panel C)—is marked. The five protomers that associate around a fivefold axis form a “pentamer.” In panels A and C, prominent surface features are labeled, namely, the flat top of one mesa (solid arrowhead), one arm of a mesa (shaded arrowhead), and one propeller blade tip (open arrowhead). Three VP1 loops form the top of the fivefold mesa, two portions of VP1 form a mesa arm, and portions of VP1 and VP2 form the blade tip. (D to G) Surface renderings of the 160S, 135S, and 80S cryo-EM reconstructions. Panels D and E show views of the 160S (top), 135S (middle), and 80S (bottom) particles along a twofold symmetry axis: stereo views of the outer surface are shown (D), as well as the inner surfaces (E). In panel E, RNA density was excised from the 160S and 135S structures with a spherical mask (r = 109 Å) so that the RNA-capsid contact regions appear smoothly spherical. Two twofold (lens shape), one threefold (triangle), and one fivefold (pentagon) symmetry axes in the central plane are shown. (F and G) Details of the outer surface viewed along the threefold (F) and fivefold (G) symmetry axes: 160S (left), 135S (middle), and 80S (right). Scale bar for panels A, D, E, F, and G = 100 Å.