Figure 4. NKX2–1GFP;AGERtdTomato reporter iPSC line enables tracking and purification of iAT1YAP5SA cells.
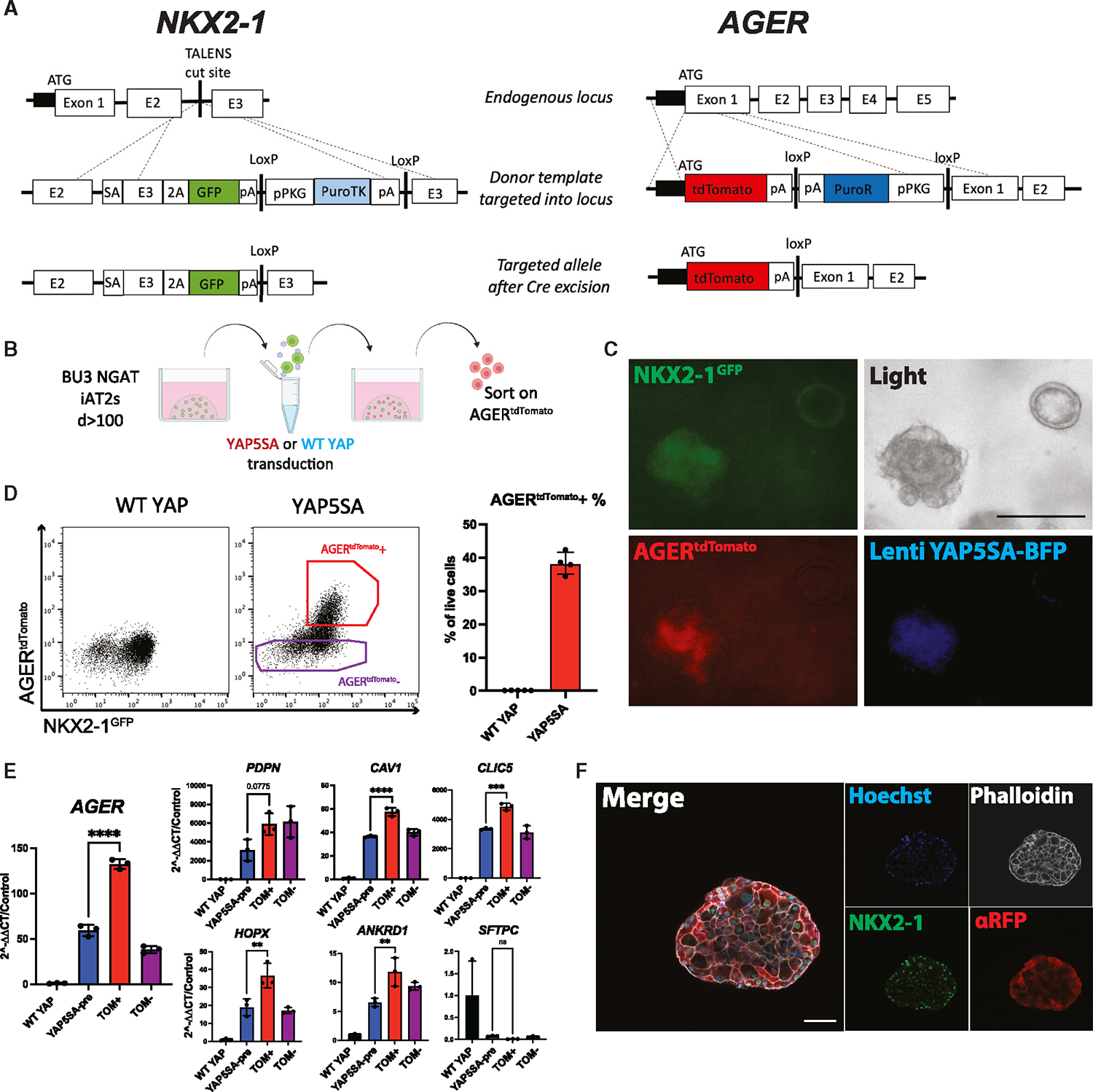
(A) Gene editing strategy to generate BU3 NKX2–1GFP;AGERtdTomato (NGAT) dual reporter iPSC line for tracking lung epithelial lineages and AT1-like cells (BU3 NKX2–1GFP reporter previously published by Hawkins et al.).54
(B) AGERtdTomato+ cells sorted and analyzed 14 days after transduction of iAT2s with lentiviral WT YAP or YAP5SA.
(C) Live cell fluorescence microscopy of YAP5SA-transduced cells growing next to an un-transduced epithelial sphere (GFP, tdTomato, and TagBFP fluorescence; scale bar, 200 μm).
(D) Flow cytometry of iAT2s (showing sorting gate for AGERtdTomato + and − cells with AGERtdTomato+ percentage quantified.
(E) Gene expression in sorted cells from (D). WT YAP, unsorted WT YAP transduced cells; YAP5SA-pre, unsorted YAP5SA transduced cells; TOM+,AGERtdTomato+ sorted cells; TOM−, AGERtdTomato−-sorted cells (N = 3 transductions, one-way ANOVA).
(F) Immunofluorescence staining of NKX2–1 protein and tdTomato (αRFP) (NKX2–1: green, F-actin: phalloidin white, tdTomato: red, nuclei: blue. Scale bars, 50 μm).
*p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.001, bars represent mean ± SD, BU3 NGAT iPSC line for all panels. See also Figure S3.
