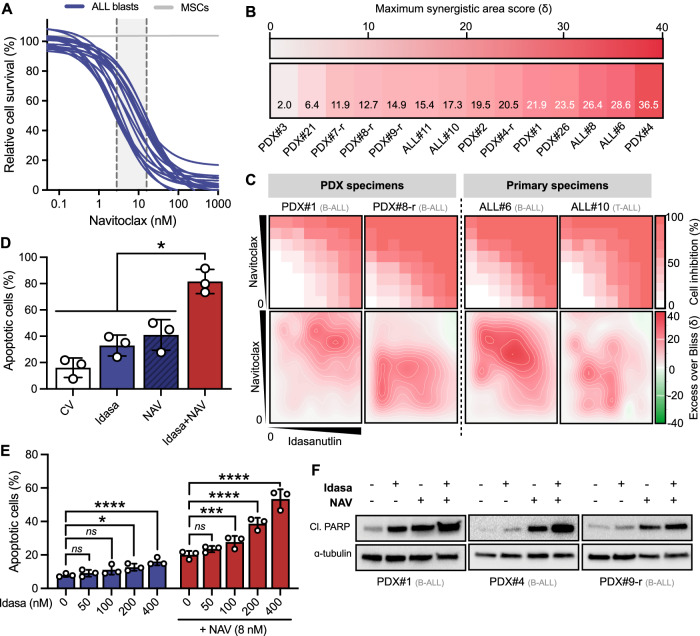
Fig. 4. Idasanutlin in combination with BCL-xL/BCL-2 inhibitor navitoclax exhibits significant ex vivo efficacy against high-risk ALL xenograft and patient samples.
A Sensitivity of primary (n = 4) and PDX (n = 13) leukemic cells exposed to increasing concentrations of NAV (0.1–1000 nM) for 96 h in ex vivo co-culture with hTERT-immortalized MSCs. Drug responses were determined by fluorescence image-based microscopy in at least technical duplicate and normalized to respective vehicle (DMSO) controls. IC50 values are based on live cell enumeration. N = 1 independent experiment in at least duplicate. The gray boundary indicates the IC50 range. B Heat map of most synergistic area Bliss synergy scores (Sarea) for idasanutlin with navitoclax combination activity in primary (n = 4) and PDX (n = 10) ALL samples ex vivo. Leukemic samples included relapse patients and patients with subtypes linked to unfavorable survival. N = 1 independent experiments in at least duplicate. C Representative dose-response matrix analyses showing cell inhibition (top) and synergistic landscape (bottom) across diverse idasanutlin-navitoclax dose combinations. D Apoptotic cells were quantified by annexin-V flow cytometry in PDX samples (n = 3) following exposure to respective IC50 concentrations of idasanutlin, navitoclax, or their combination for 48 h. Error bars show mean ± s.d. Combination treated cells were compared to respective single drugs by paired t-tests. E Apoptotic cells were quantified by annexin-V flow cytometry in PDX#4 cells following exposure to indicated concentrations of idasanutlin ± navitoclax (8 nM) for 24 h (n = 3). Error bars show mean ± s.d. Data were analyzed by two-way ANOVA with Tukey posttest for multiple comparisons. F Immunoblots showing induction of cleaved PARP (Asp214) in PDX samples (n = 3) following exposure to respective IC50 concentrations of idasanutlin, navitoclax, or their combination for 24 h. ɑ-tubulin served as the loading control. ns=not significant, *p value < 0.05, ***p value < 0.001, ****p value < 0.0001.