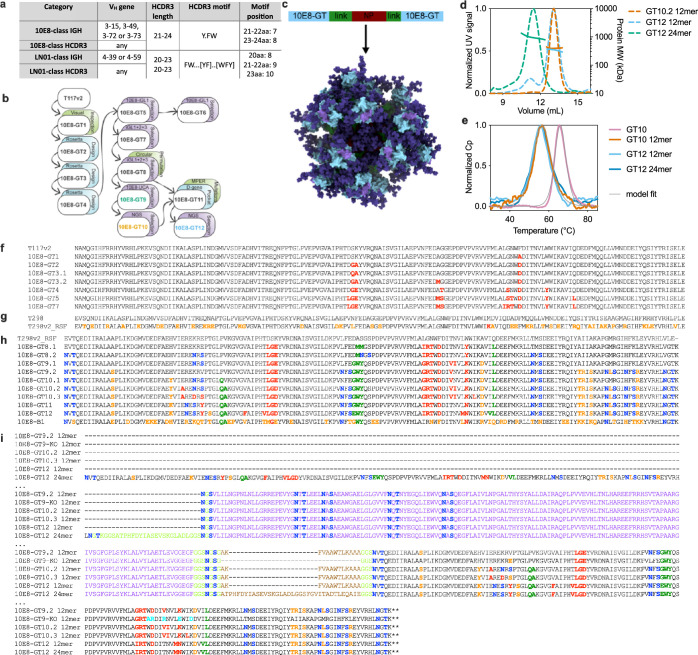
Extended Data Fig. 1. Design and properties of immunogens.
a, Overview of 10E8-class and LN01-class antibody categories. HCDR3 motifs are shown as regular expressions that were used to query the database. If multiple amino acids were allowed at the same position, they are shown in square brackets; positions in which all amino acids were allowed are indicated as ‘.’. b, Schematic of the development of MPER-GT scaffolds. c, Schematic overview of nanoparticle formation by genetic fusion of the immunogen (T2983−GT) to each terminus of the 3-dehydroquinase nanoparticle from Thermus thermophilus (NP) via flexible linkers containing exogeneous T-help peptides derived from Aquifex aeolicus lumazine synthase (link). The epitope scaffold is shown in light blue, the MPER graft in purple, the linker in green, the nanoparticle in red and glycans in dark blue. d, SEC-MALS traces of 10E8-GT NPs. Normalized UV280 absorptions are shown as dotted lines and protein molecular weights of main peaks are shown as solid lines. e, DSC measurements of the indicated monomers and nanoparticles with results from a fit indicated in light grey. f, Amino acid sequences of 10E8-GT epitope scaffolds through generation 7, none of which had the circular permutation present in later generations. Germline-targeting mutations are highlighted in red; N-linked glycosylation sites are blue; the D-gene binding pocket is green; resurfacing and/or solubility enhancing mutations are orange. All sequences are succeeded by a 6x His-tag, unless the protein ends in stop codons (denoted by symbol *). 10E8-GT8.2 through 10E8-GT12 are preceded by a mammalian secretion signal. g, Resurfaced T298v2 sequences compared to previously published T298 32. Colors as in f. h, Amino acid sequences of monomeric immunogens based on resurfaced circularly permutated T298v2, with colors as in f. i, Amino acid sequences of multivalent nanoparticles. Sequences are wrapped over multiple lines. The 3-dehydroquinase is shown in purple; additional T help epitopes are brown; epitope KO mutations are cyan; and all other colors are shown as in f.