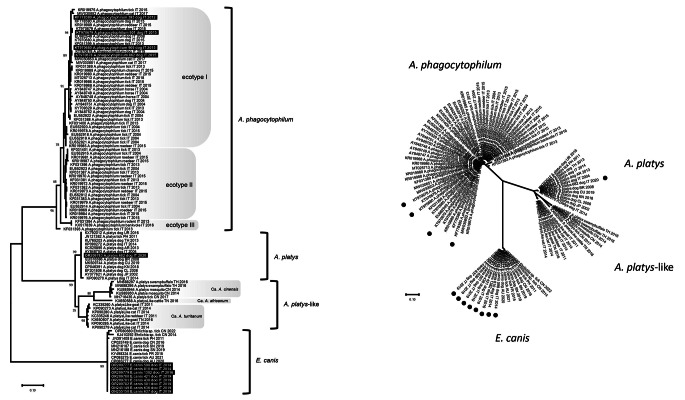
Fig. 2.
Phylogenetic tree based on partial heat shock gene (groEL) nucleotide sequences of Anaplasma phagocytophilum, A. platys and Ehrlichia canis. Phylogeny was evaluated using the Maximum Likelihood method and the Tamura 3-parameter model implemented on MEGA 11 software version 11.0.11 on multiple alignment constructed with nucleotide sequences obtained in this study, 55 reference sequences of A. phagocytophilum, 17 of A. platys, 8 of A. platys-like and 9 of E. canis retrieved from GenBank. On the left a traditional rectangular branch style of the tree and on the right a radiation branch style of the tree. Identification of the sequences undergoes the following nomenclature: GenBank accession number, species, strain (only for sequences obtained in this study), host, country (AR: Argentina, AU: Australia, BR: Brazil, CL: Chile, CN: China, CU: Cuba, FR: France, IT: Italy, JP: Japan, KN: Saint Kitts and Navis, PH: Philippines, SN: Senegal, TH: Thailand, TN: Tunisia, UR: Uruguay), and collection date (or date of database submission). Statistical support was provided by bootstrapping with 1000 replicates and values indicated on the respective branches. Highlighted in black or black circles: sequences generated in this study. Main clusters of A. phagocytophilum are labelled following the ecotype classification proposed by Jahfari and colleagues (Jahfari et al. 2014)