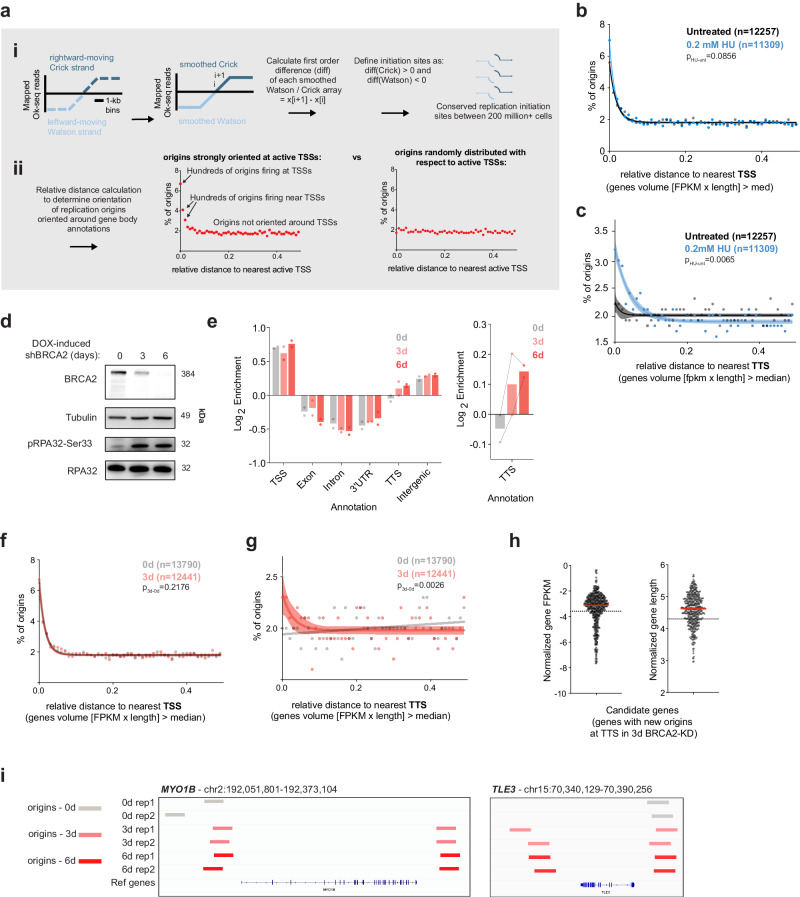
Fig. 1. Loss of BRCA2 causes dormant origin firing at TTSs of long, highly transcribed genes.
a Identification of new origins at TTSs upon replication stress and schematic of Okazaki-fragment sequencing origin analysis. (i) 1 kb-binned Crick and Watson Ok-seq reads are Hann smoothed and replication origins calls are defined as regions of increasing Crick-strand derivative and decreasing Watson-strand derivative from smoothed reads (see “Methods” section). (ii) Relative distance calculation orients origins to genomic annotations (TSS, TTS). Replication origins are prevalent at active TSSs indicated by a high proportion of origins at low relative distances in active TSSs (left). If origins were randomly distributed with respect to active TSSs, there would be an equal proportion at all relative distances (right). b Relative distance origin analysis of previously published untreated and HU-treated RPE-1 cells (Chen et al.27) relative to TSSs of transcriptionally high-volume genes (FPKM × length > median). N = # of origins per condition. Percent of origins at each relative distance were fit to a nonlinear regression curve (solid line) with a 90% confidence interval (shaded). p-values calculated using a one-sided, paired t-test between conditions for the first three relative distance bins (0, 0.01, 0.02) (see “Methods” section). c Relative distance origin analysis of untreated and HU-treated RPE-1 cells relative to TTSs of high-volume genes. The solid line represents origins at each relative distance fit to a nonlinear regression curve with a 90% confidence interval (shading). p-values calculated using a one-sided, paired t-test between conditions for the first three relative distance bins (0, 0.01, 0.02). d Whole cell lysates of immortalized fallopian tube epithelial cells (FTEs) with doxycycline (DOX)-inducible BRCA2-shRNA after 0, 3, and 6 days of treatment were analyzed by western blot for validation of knockdown (KD) and pRPA-related replication stress. Tubulin is a loading control for this blot and subsequent western blots. Results reproducible for at least 3 biological replicates. e (Right) Log2 annotation enrichment of FTE Ok-seq origins at 0, 3, and 6 days of BRCA2-KD. N = 2 biological Ok-seq replicates each (left). Zoomed TTS annotation (right); lines connect biological replicates. f Relative distance origin analysis of untreated and BRCA2-KD FTEs relative to TSSs of high-volume genes. For all subsequent experiments, 3 days of dox treatment was used for BRCA2-KD. The solid line represents origins at each relative distance fit to a nonlinear regression curve with a 90% confidence interval (shading). p-values calculated using a one-sided, paired t-test between conditions for the first three relative distance bins (0, 0.01, 0.02). g Relative distance origin analysis of untreated and BRCA2-KD FTEs relative to TTSs of high-volume genes. The solid line represents origins at each relative distance fit to a nonlinear regression curve with a 90% confidence interval (shading). p-values calculated using a one-sided, paired t-test between conditions for the first three relative distance bins (0, 0.01, 0.02). h Log10 normalized gene FPKM and normalized gene length of candidate genes with new origins at TTSs with BRCA2-KD (n = 418 genes). The dotted line represents the average FPKM and gene length of all genes (N = 18250) from RNA-seq analysis of FTEs. i Two representative candidate genes with new origins at TTSs with BRCA2-KD in two Ok-seq biological replicates each with 0, 3, and 6 days of knockdown: MYO1B and TLE3. Source data are provided as a Source Data file.