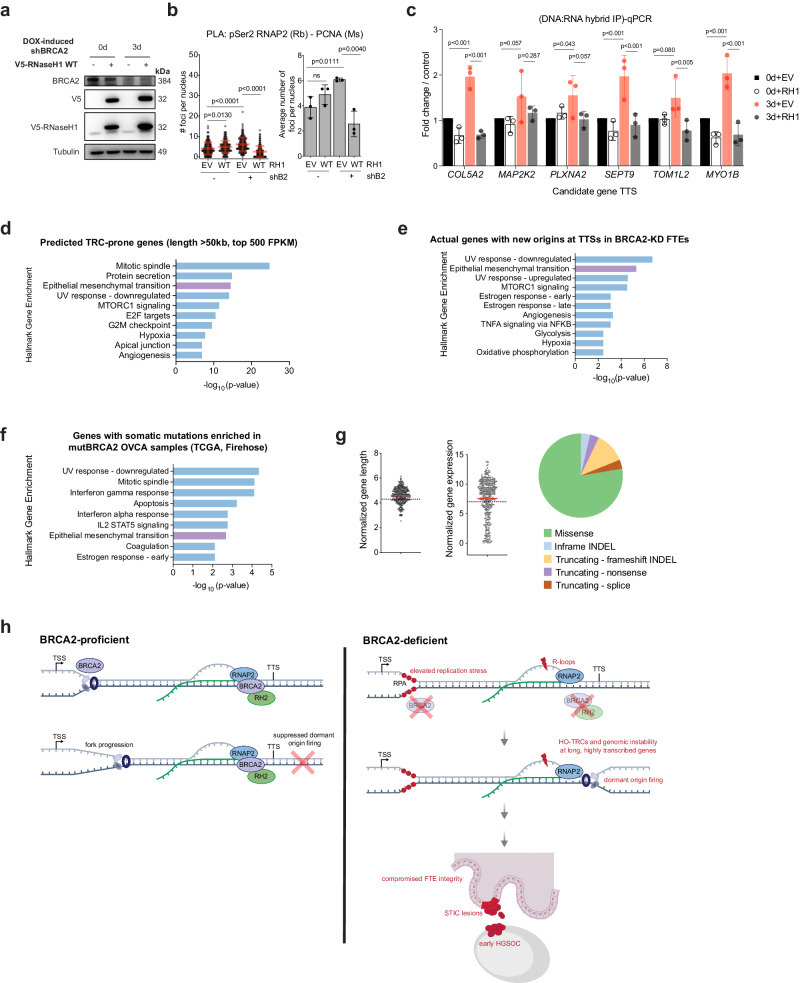
Fig. 5. HO-TRCs are sites of genomic instability in BRCA2-deficient cells.
a Whole cell lysates of FTEs were analyzed by western blot. Empty vector or V5-tagged RNase H1 (RH1) plasmids were transiently transfected for 24 h prior to collection in WT- or KD-BRCA2 FTEs. Results reproducible for at least 3 biological replicates. b PLA of pSer2-RNAP2 and PCNA of WT- and KD-BRCA2 ± RH1 overexpression. The data presented shows at least 150 nuclei from 3 biological replicates. p-values calculated using unpaired two-tailed t-tests between conditions. Left: number of foci per nucleus. Right: average number of foci per nucleus per biological replicates. Error bars = mean, std. c DRIP-qPCR analysis of 6 candidate gene TTSs in WT- and KD-BRCA2 FTEs ± RH1 overexpression. The bar graph shows % of input normalized to the control sample (0d + EV) of 3 biological replicates. p-values calculated using two-way ANOVA with Tukey’s multiple comparisons test. Error bars = mean, std. d Hallmark gene set analysis (GSEA) of genes length >50 kb and highly expressed (top 500 FPKM) from RNA-seq of FTEs. Top hallmark gene sets are shown as -log10(p-value) p-values calculated using the hypergeometric distribution of overlapping genes over all genes in the gene universe. e Hallmark GSEA of candidate genes with new origins at TTSs in shBRCA2-KD based on Ok-seq origin analysis. p-values calculated using the hypergeometric distribution of overlapping genes over all genes in the gene universe. f Hallmark GSEA of genes with mutations enriched in mutBRCA2 samples vs all mutTP53 samples in ovarian serous cystadenocarcinoma patients (TCGA Firehose study). p-values calculated using the hypergeometric distribution of overlapping genes over all genes in the gene universe. g Characterization of genes with mutations enriched in mutBRCA2 samples vs all mutTP53 samples in ovarian serous cystadenocarcinoma patients (N = 376) (TCGA Firehose study): log normalized gene length (left); log normalized gene expression (RNA-seq V2 RSEM) (middle); mutation type (right). h Schematic model of HO-TRCs at TTSs in an early BRCA2-deficient HGSOC. Left: BRCA2 protects stalled replication forks to promote fork progression of canonical forks at TSSs. BRCA2 promotes transcription elongation and R-loop resolution through RNase H2 recruitment. Right: in BRCA2 deficiency, increased replication stress and transcriptional dysregulation. Dormant origin firing at TTSs of long, highly transcribed genes causes HO-TRCs with elongating RNAP2 and DNA damage. TRC-prone genes maintain fallopian tube epithelial (FTE) integrity, which is compromised in early BRCA2-deficient HGSOC lesions. Source data are provided as a Source Data file.