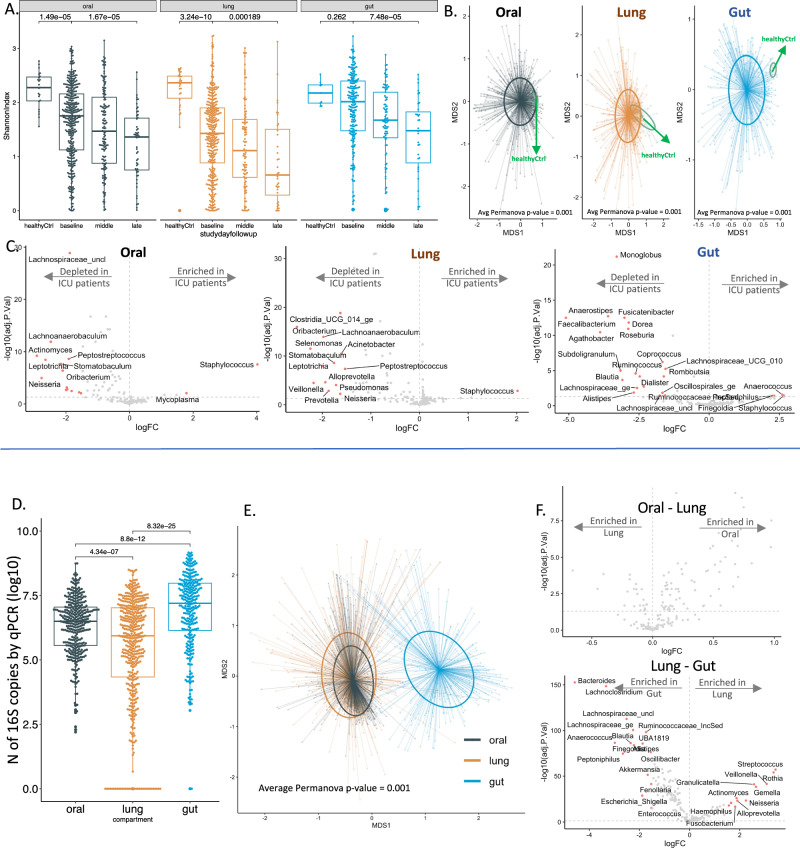
Fig. 1. Intra- and inter-compartment comparisons of microbiota profiles by Illumina 16S-Seq reveal features of dysbiosis in all three body compartments in critically ill patients. Panels A–C: Intra-compartment comparisons between ICU patients and healthy controls.
A Samples from critically ill patients had significantly lower alpha diversity (Shannon index obtained post-rarefication with random subsampling of reads in samples with ≥1000 16S rRNA gene reads) compared to corresponding healthy control samples in each compartment (Wilcoxon test p < 0.001), with a further decline of Shannon index over time in longitudinal samples in critically ill patients (Wilcoxon test p < 0.001). B Baseline samples from critically ill patients had markedly significant differences in beta diversity (Bray-Curtis indices in centered-log ratio transformed [CLR] abundances following random subsampling of reads in samples with ≥ 1000 reads) compared to healthy controls (visualized with Principal Coordinates Analysis [PCoA] and statistically compared with permutational analysis of variance [permanova] p values < 0.001, adjusted for multiple comparisons with the Bonferroni method). C Taxonomic composition comparisons with the limma package showed high effect sizes and significance thresholds (threshold of log2-fold-change [logFC] of CLR-transformed abundances >1.5; Benjamini-Hochberg adjusted p value < 0.05), revealing depletion for multiple commensal taxa in critically ill patients samples, with significant enrichment for Staphylococcus in oral and lung samples, and Anaerococcus and Staphylococcus in gut samples (significant taxa shown in red in the volcano plots). Panels D–F: Inter-compartment comparisons among ICU patients. D Lung samples had lower bacterial burden compared to oral and gut samples by 16S rRNA gene qPCR (all Wilcoxon test p < 0.001). E PCoA plot of beta-diversity shows compositional similarity for the oral and lung compartments, which were compositionally dissimilar to gut samples (permanova p < 0.001). F Taxonomic comparisons between compartments revealed that no specific taxa were systematically different between oral and lung microbiota above the threshold of logFC≥1.5, whereas in gut-lung comparisons, lung communities were enriched for typical respiratory commensals (e.g. Rothia, Veillonella, Streptococcus) and gut communities for gut commensals (e.g. Bacteroides, Lachnoclostridium, Lachnospiraceae). Source data are provided as a Source Data file. Displayed data include 583 oral, 543 lung, and 343 gut samples from ICU patients, as well as 23 oral, 32 lung, and 7 gut samples from healthy controls. Data displayed as boxplots with individual dots have their median as the line inside the box, interquartile range (25th–75th percentile) as the box itself, whiskers extend to 1.5 times the interquartile range, and individual dots beyond whiskers signify outlier observations. All statistical tests were two-sided.