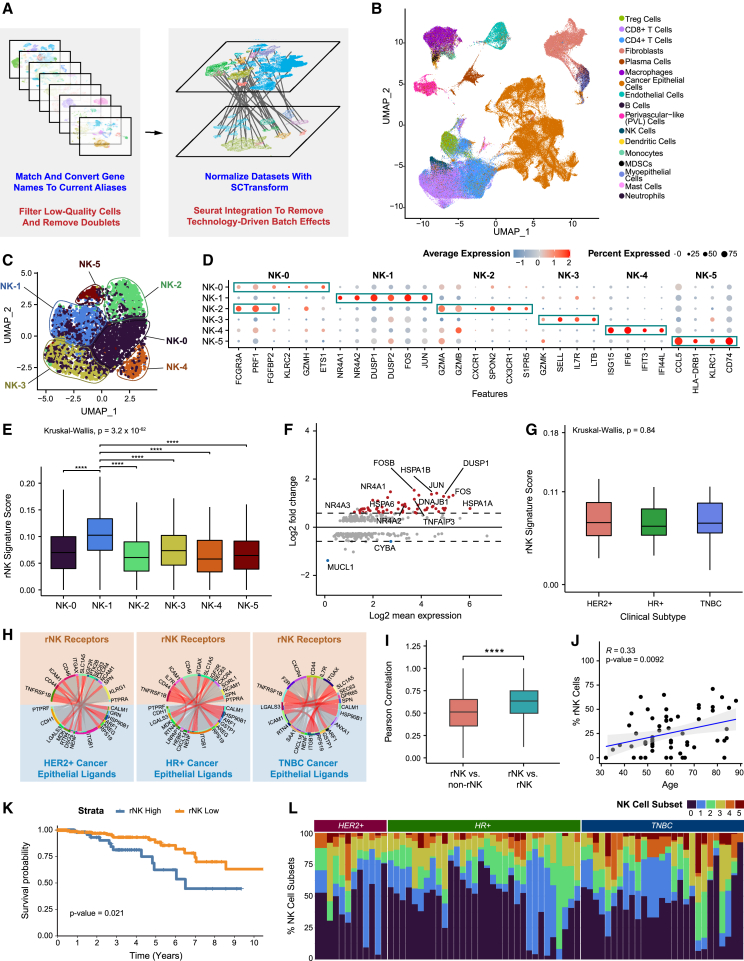
Figure 1.
Integrated scRNA-seq dataset of primary breast cancer identifies six NK cell subsets in breast cancer
(A) Brief overview of the processing and integration pipeline for 8 primary breast cancer datasets.
(B) UMAP visualization of 236,363 cells across 119 samples from 88 patients analyzed by scRNA-seq.
(C) UMAP visualization showing major subsets of natural killer (NK) cells.
(D) Bubble heatmap showing expression of upregulated differentially expressed genes for each major NK cell subset (Bonferroni-adjusted p < 0.05).
(E) Boxplot showing expression of the rNK cell signature in each NK cell subset. NK-1 was significantly different from all other clusters (Kruskal-Wallis p < 0.0001, with post hoc Dunn test p values shown; ∗∗∗∗p < 0.0001).
(F) MA plot of differentially expressed genes between rNK and non-rNK cells (Bonferroni-adjusted p < 0.05).
(G) Boxplot showing the expression level of the rNK signature by clinical subtype. No significant difference was found between subtypes (Kruskal-Wallis p > 0.05).
(H) Circos plots showing representative predictive receptor-ligand pairs between rNK cells and all cancer epithelial cells separated by clinical subtype. Shared receptors across all subtypes are colored in red.
(I) Boxplot showing the Pearson correlations of rNK signature gene expression in reprogrammed NK (rNK) cells compared with non-rNK cells versus rNK cells compared with rNK cells (across all clinical subtypes of breast cancer). Pearson correlations between rNK cells and rNK cells are higher than those between rNK cells and non-rNK cells (two-sided Wilcoxon test, ∗∗∗∗p < 0.0001).
(J) Scatterplot showing the Pearson correlation of age and proportion of rNK cells by sample (p <0.01).
(K) Kaplan-Meier plot showing worse clinical outcome in breast cancer patients with high expression of the rNK cell gene signature (log rank test, p < 0.05).
(L) Bar plot showing relative proportions of NK subsets across tumor samples and clinical subtypes.
See also Figures S1–S5 and Data S1 and S4.