Figure 3.
Cancer epithelial cell heterogeneity can be defined by 10 GEs that influence immune cell interactions
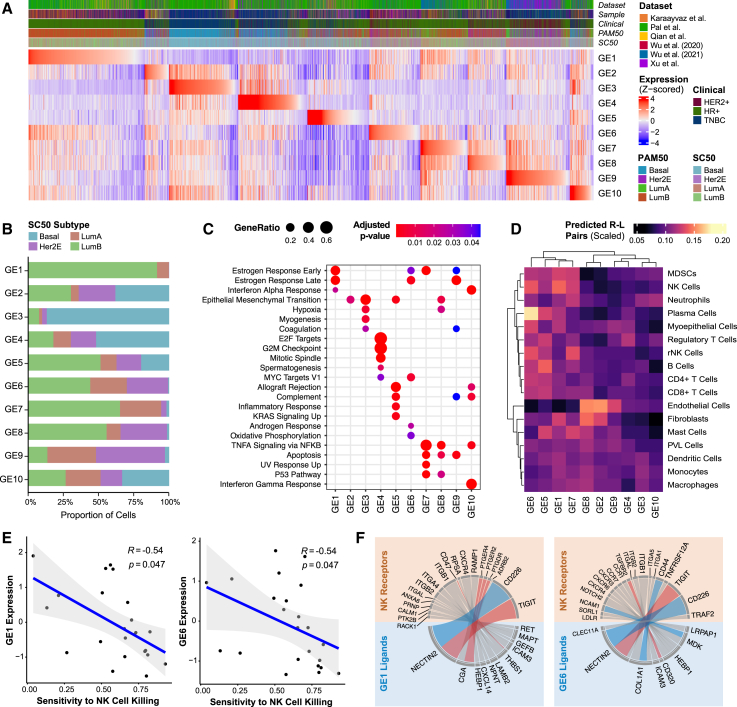
(A) Heatmap of Z-scored signature scores of the 10 identified gene elements (GEs) representing all cancer epithelial cells, ordered based on the maximum Z-scored GE signature score. Annotations represent dataset origin, clinical subtype, PAM50 subtype, and SC50 subtype. The “sample” annotation was included to demonstrate that no individual patient sample contributed heavily to a particular GE.
(B) Percentage of cancer epithelial cells assigned to each GE by molecular subtype.
(C) Gene set enrichment using ClusterProfiler of the differentially expressed genes by GE. Significantly enriched gene sets from the MSigDB Hallmark collection are shown (Benjamini-Hochberg-adjusted p < 0.05).
(D) Heatmap of the scaled number of curated predicted receptor-ligand pairs between cancer epithelial cells by GE and interacting immune and stromal cells.
(E) Scatterplots showing Spearman correlations of expression of NK-cell related GE1 and GE6 with sensitivity to NK cell killing (Benjamini-Hochberg-adjusted p < 0.05).
(F) Circos plots showing curated receptor-ligand pairs between cancer epithelial cells that highly express NK cell-related GE1 and GE6 with NK cells. NK cell activating receptor-ligand pairs are colored blue; NK cell inactivating receptor-ligand pairs are colored red.
See also Figure S8.