Figure 4.
GE-immune interactions predict response to anti-PD-1 therapy
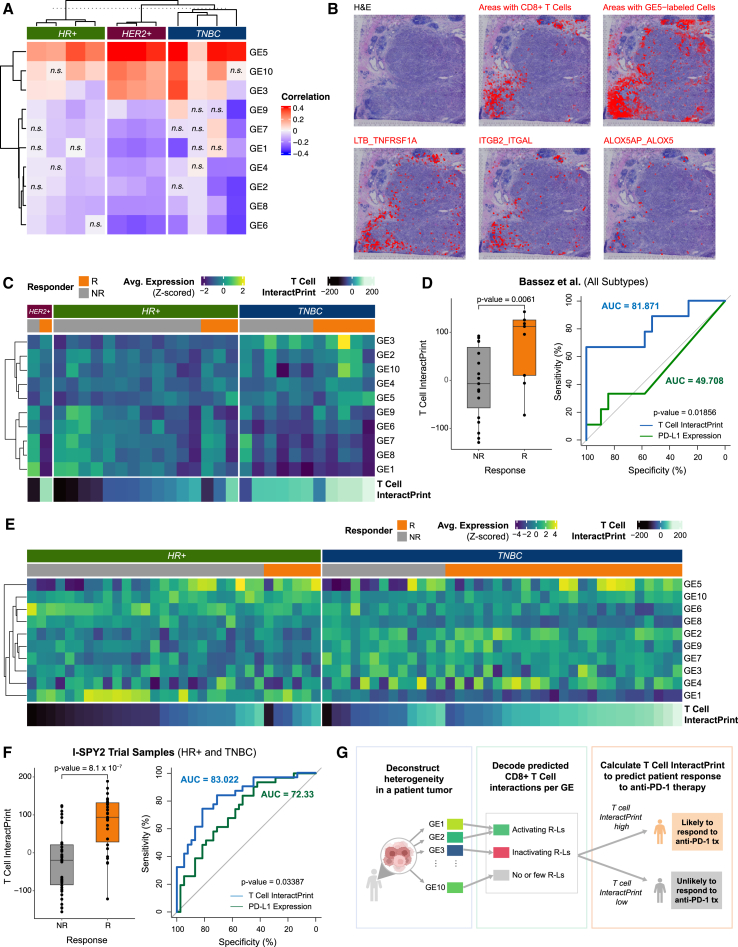
(A) Heatmap of Pearson correlations between expression of each of the 10 GEs and the presence of CD8+ T cells for 6 spatial transcriptomics samples across spots containing CD8+ T cells (n.s., Benjamini-Hochberg-adjusted p > 0.05).
(B) For a representative TNBC sample, pathological annotation of morphological regions into distinct categories. UCell signature scores of CD8+ T cells are overlaid onto spatial tumor sample spots (red). A UCell signature score of GE5 (a CD8+ T cell activating GE) is overlaid onto tumor sample spots (red). A colocalization score for ITGB2_ITGAL, LTB_TNFRSF1A, and ALOX5AP_ALOX5 (predicted receptor-ligand pairs for GE5 and CD8+ T cells) is overlaid onto tumor sample spots (red).
(C) Heatmap of average expression of each of the 10 GEs across cancer epithelial cells in each sample from Bassez et al.57 T cell InteractPrint is shown below.
(D) Boxplot showing T cell InteractPrint prediction of response to anti-PD-1 therapy across all clinical subtypes in Bassez et al.57 (R, responder; NR, non-responder; p < 0.05). Also shown is the AUC of ROC comparing the performance of T cell InteractPrint (AUC = 81.87) and of PD-L1 expression (AUC = 49.71) in Bassez et al.57 samples (bootstrap test with n = 10,000, p < 0.05).
(E) Heatmap of average expression of each of the 10 GEs across cancer epithelial cells in each sample from the I-SPY2 trial. T cell InteractPrint is shown below.
(F) Boxplot showing T cell InteractPrint prediction of response to anti-PD-1 therapy across all clinical subtypes in I-SPY2 trial samples (two-sided Wilcoxon test p <0.0001). Also shown is the AUC of ROC comparing the performance of T cell InteractPrint (AUC = 83.02) and of PD-L1 expression (AUC = 72.33) in the I-SPY2 trial (bootstrap test with n = 10,000, p <0.05).
(G) Schematic of T cell InteractPrint to predict patient response to anti-PD-1 therapy.
See also Figure S9.