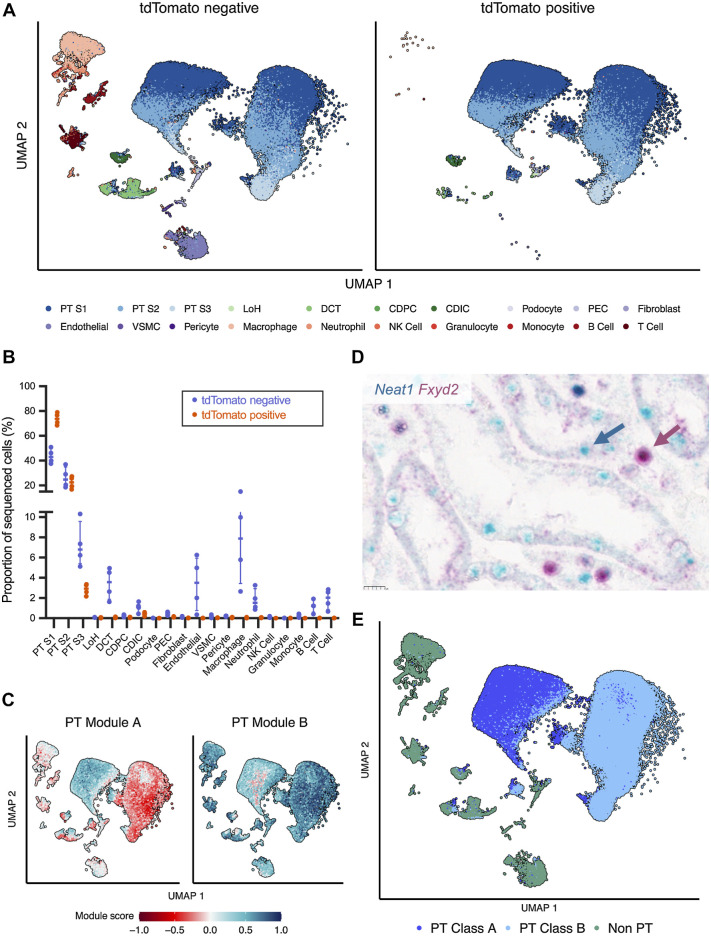
Figure 2.
scRNA-seq on flow-sorted renal cells. A, UMAP plot of tdTomato-negative (left) or tdTomato-positive cells (right) from kidneys of Control mice harvested at the early time point. Cells are colored by inferred cell type. LoH, loop of Henle; DCT, distal convoluted tubule; CD, collecting duct; PC, principal cell; IC, intercalated; PEC, parietal epithelial cell; VSMC, vascular smooth muscle cell; NK, natural killer cell. B, Proportion of sequenced cells inferred to be of each cell type in tdTomato-positive or tdTomato-negative populations from kidneys of Control mice harvested at the early time point. Median and interquartile range plotted. C, UMAP plot depicting expression of PT Module A (left) and PT Module B (right) genes in PT cells from Control mice. D, Representative in situ RNA hybridization exhibiting spatially distinct expression of Neat1 (blue) and Fxyd2 (red) mRNA in FFPE kidney cortex from Control mice harvested at the early time point. Scale bar, 10 μm. Magnification, ×40. E, UMAP plot depicting cells from Control mice at the early time point. Cells are colored by assigned PT Class. A–E, scRNA-seq data shown for n = 3 female (3F) and n = 1 male (1M) Control mice.