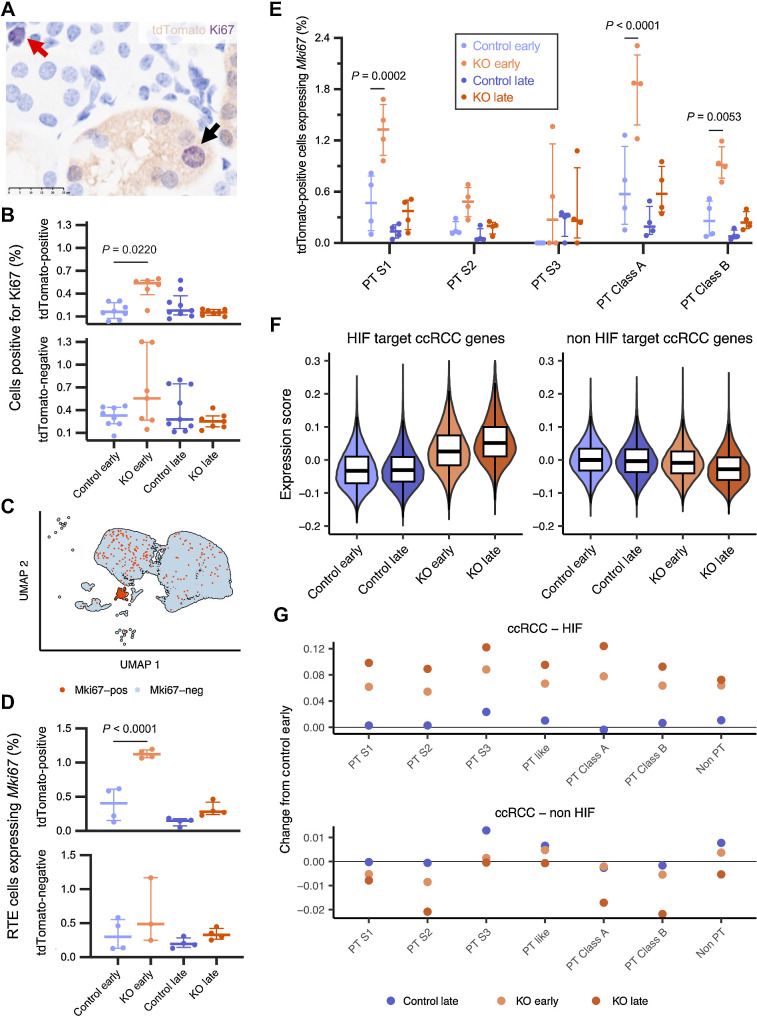
Figure 6.
Vhl-null cells exhibit time-dependent proliferation and association with ccRCC-like gene expression. A, Representative dual IHC for tdTomato (brown) and Ki67 (purple) counterstained with hematoxylin in kidneys of KO mice harvested early after recombination. Scale bar, 25 μm. Magnification, ×40. Black arrow, dual-positive cell; red arrow, tdTomato-negative Ki67-positive cell. B, Proportion of tdTomato-positive (top) or tdTomato-negative (bottom) cells that are positive for Ki67 by dual IHC in kidneys of Control and KO mice harvested early and late after recombination (n = 2F, 6M for Control early; n = 4F, 2M for KO early; n = 4F, 5M for Control late; n = 1F, 6M for KO late). Pairwise comparisons by Kruskal–Wallis test with Dunn correction. C, UMAP plot depicting RTE cells from Control and KO mice at the early and late time points. Orange, cells expressing Mki67. D, Proportion of tdTomato-positive (top) or tdTomato-negative (bottom) RTE cells that express Mki67 in different conditions. Pairwise comparisons tested by one-way ANOVA with Holm–Šídák correction. E, Proportion of tdTomato-positive cells of different PT identities that express Mki67 in different conditions. Pairwise comparisons tested by two-way ANOVA with Holm–Šídák correction. F, Violin plot overlaid with boxplot depicting expression score for genes upregulated in ccRCC cells known to be HIF targets (left) and not known to be HIF targets (right) in tdTomato-positive cells from Control and KO mice harvested at early or late time points. G, Scatter plot depicting changes in mean expression scores for HIF-target (top) and non-HIF-target (bottom) genes specifically upregulated in ccRCC, in tdTomato-positive cells of different PT identities from different conditions when compared with those from Control mice at the early time point. B, D, and E, Median and interquartile range plotted. Only significant (P < 0.05) comparisons shown. C–G, scRNA-seq data shown for n = 3F, 1M mice for tdTomato-positive and tdTomato-negative Control early and Control late samples; n = 2F, 1M mice for tdTomato-negative KO early samples; n = 2F, 2M mice for tdTomato-positive KO early samples; n = 2F, 2M mice for tdTomato-positive and tdTomato-negative KO late samples.