Figure 1.
Schematic of genome-first assessment of pathogenic/likely pathogenic or predicted loss-of-function (P/LP/LoF) variants in the BioMe Biobank at Mount Sinai and UK Biobank
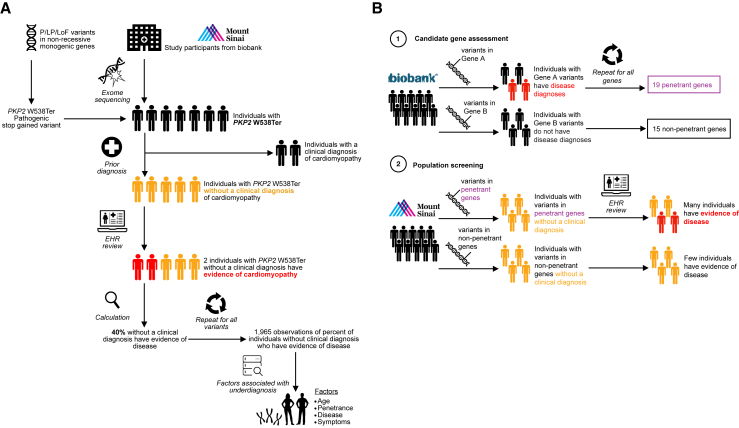
(A) Study design for genome-first evaluation in the BioMe Biobank at Mount Sinai using the plakophilin 2 (PKP2) W538Ter variant as an example. A total of P/LP/LoF variants in non-recessive monogenic genes for nine genetic disorders were curated: (1) variants reported as P/LP in the ClinVar repository with a minimum review status of two (multiple submitters) and previously unreported LoF variants identified by Variant Effect Predictor were obtained, and (2) non-recessive genes with a monogenic disease predisposition were identified from Online Mendelian Inheritance in Man and corroborated with literature review. A total of 29,039 participants with exome sequence and electronic health record (EHR) data were included in the study. This yielded 644 observations of 303 P/LP/LoF variants in 614 individuals. As an example, PKP2 W538Ter was identified in seven individuals, of whom two had a prior clinical diagnosis of cardiomyopathy in the EHR. The remaining five individuals lacked a clinical diagnosis, of whom two (40%) were discovered to have EHR evidence of cardiomyopathy. This procedure was repeated for all variants to produce a dataset of the percentage of clinically undiagnosed observations that had evidence of disease, which was used to assess factors associated with the presence of disease in clinically undiagnosed individuals. Factors comprised the gene containing the variant, age of individuals, disease, and symptoms.
(B) Evaluation of clinically undiagnosed but symptomatic individuals with P/LP/LoF variants in BioMe Biobank by gene penetrance observed in UK Biobank. A total of 34 target genes were identified in an independent cohort from UK Biobank, for which disease was either observed in individuals with P/LP/LoF variants in the gene (19 penetrant genes) or not observed (15 non-penetrant genes). The proportion of clinically undiagnosed observations with disease evidence in BioMe Biobank was then compared between the penetrant genes and non-penetrant genes.