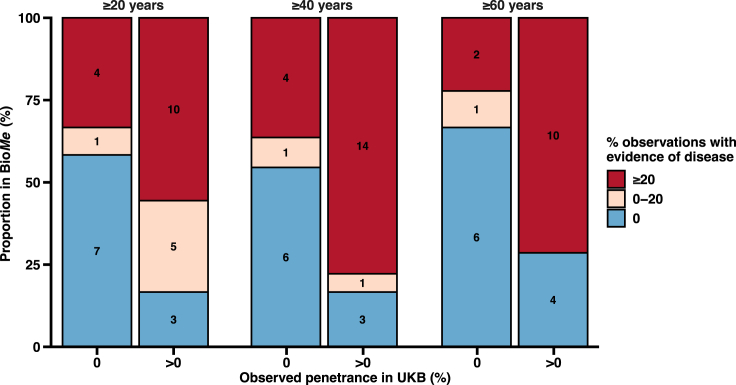
Figure 3.
Rates of disease evidence found in clinically undiagnosed individuals with pathogenic/likely pathogenic or predicted LoF (P/LP/LoF) variants in BioMe Biobank (BioMe) for genes with versus without disease occurrence in UK Biobank (UKB)
A subset of 34 genes and six diseases that had clinically undiagnosed observations in BioMe were identified in UKB. Disease was observed in individuals from UKB with P/LP/LoF variants in 19 genes (penetrant genes) and not observed for 15 genes (non-penetrant genes) (STAR Methods). Yield of disease evidence in BioMe was then compared for P/LP/LoF variants found in penetrant genes versus non-penetrant genes. For each gene, clinically undiagnosed observations were evaluated for electronic health record evidence of symptomatic disease, and the proportion of observations with disease evidence was categorized as 0% (blue), between 0% and 20% (tan), or ≥20% (red). This analysis was completed three times using individuals who were ≥20, ≥40, and ≥60 years of age. For example, for individuals ≥40 years of age, 14 of 18 (78%) genes with disease observed in UKB had ≥20% observations with disease evidence compared to 4 of 11 (36%) genes with no disease observed in UKB.