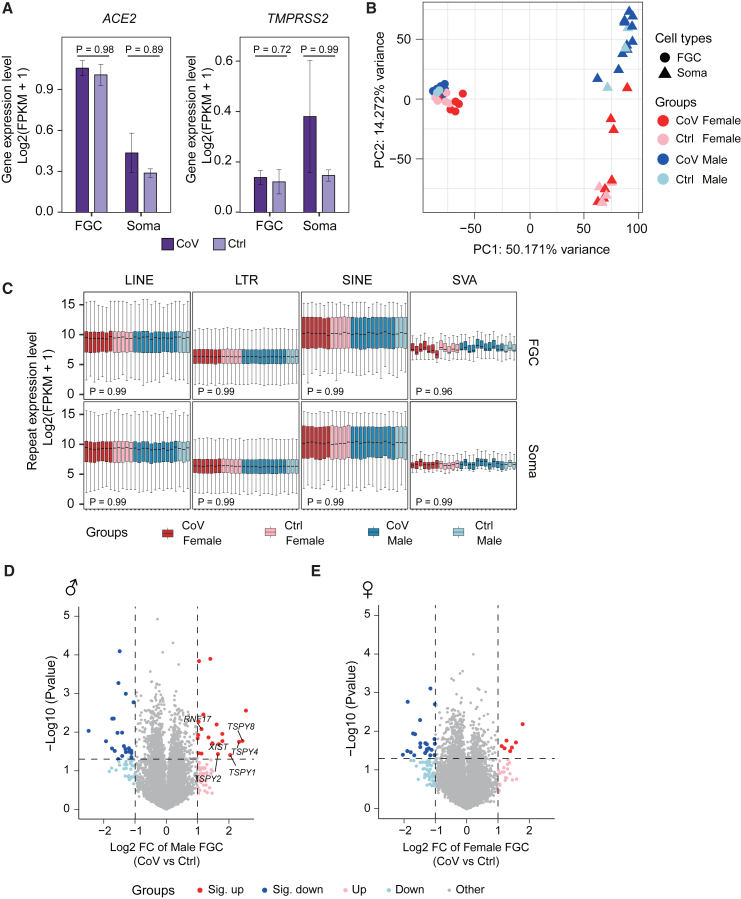
Figure 2.
Transcriptome reveals limited impact on human FGC development following maternal SARS-CoV-2 infection
(A) Expression level of selected SCARFs (SARS-CoV-2- and coronavirus-associated receptors and factors) in FGCs and gonadal somatic cells. The expression level is represented as log2(FPKM+1). The p values were calculated by Wilcoxon rank-sum test. CoV_n = 18; Ctrl_n = 8.
(B) Principal-component analysis (PCA) of the transcriptomes of FGCs and gonadal somatic cells from the CoV and Ctrl groups. Circle, FGC; triangle, gonadal somatic cell; red color scheme, female; blue color scheme, male. The variance of PC1 and PC2 is shown.
(C) Expression level of major human repetitive element classes in FGCs and gonadal somatic cells. The expression level is represented as log2(FPKM+1). Wilcoxon rank-sum test was performed to examine the statistical difference between the CoV group and Ctrl group. Red color scheme, female; blue color scheme, male. The boxplots are ordered by gestational age at termination, horizontally in each group.
(D and E) Volcano plots of differentially expressed genes (DEGs) identified by CoV group versus Ctrl group in male (D) or female (E) FGCs. The alternation of gene expression is represented as log2 fold change (log2 FC). Dark red and dark blue dots indicate genes significantly upregulated or downregulated in the CoV group, respectively (|log2 FC| > 1, p < 0.05; male_sig._up_n = 20, male_sig._down_n = 23, female_sig._up_n = 7, female_sig._down_n = 23). Light red and light blue dots indicate genes upregulated or downregulated in the CoV group (|log2 FC| > 1, p ≥ 0.05; male_up_n = 37, male_down_n = 30, female_up_n = 20, female_down_n = 50). Gray dots indicate other genes. The p values were calculated by Student’s t test. CoV_Male_n = 11, Ctrl_Male_n = 3, CoV_Female_n = 7, Ctrl_Female_n = 5.
See also Figures S2 and S3.