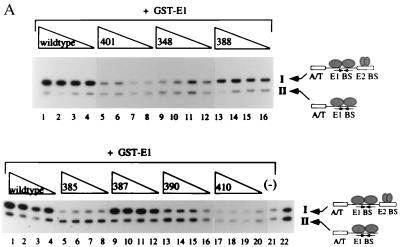
FIG. 2.
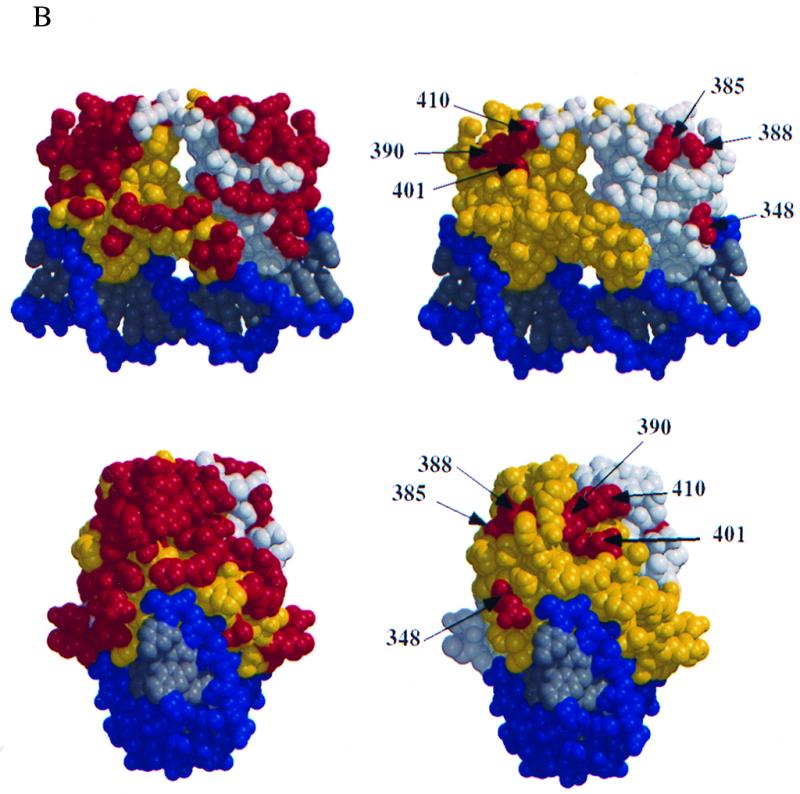
(A) Six mutant proteins are defective for cooperative binding with E1. Two different probes, one containing the BPV minimal ori with a high-affinity E2 binding site (I) and one containing an E1 binding site alone (II), were incubated with 6 ng of GST-E1 alone (lane 21, bottom) or with either the wt E2 DBD (lanes 1 to 4, top and bottom), or seven mutant E2 DBDs (lanes 5 to 16, top, and lanes 5 to 20, bottom). Equal quantities of wt and mutant proteins, based on Western blot analysis, were used in four twofold titrations. Probes bound by GST-E1 were recovered with glutathione-agarose beads and analyzed on a 6% urea gel. E2-dependent stimulation of binding was measured by comparing the amounts of probes II and I recovered. (B) Five mutations which do not affect DNA binding form two distinct patches on the E2 DBD. Shown are two different views of a space-filling model of the E2 DBD bound to its cognate binding site, created by the BOBSCRIPT program and Raster3D (6, 10, 17). In the images on the right, all mutations which were made on the surface of the E2 DBD are in red. The images on the left show the same two views with mutations that affect the ability of the E2 DBD to interact with E1 in red.