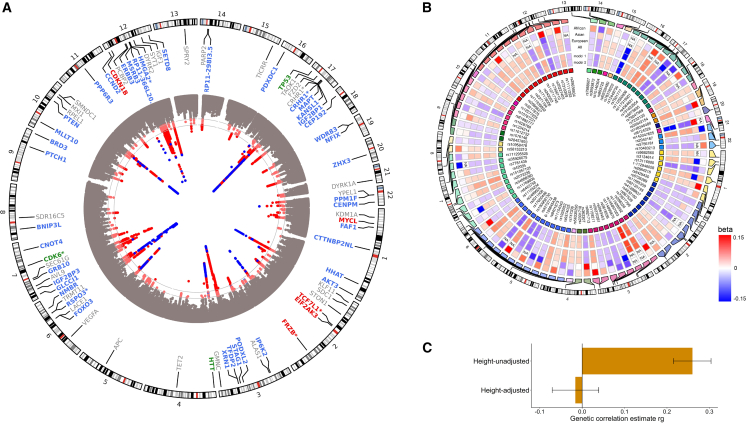
Figure 1.
Genome-wide association studies on human head size
(A) Circos Manhattan plot of the European ancestry head size GWAS, with gray lines corresponding to genome-wide significant (p < 5 × 10−8) or sub-significant (p < 1 × 10−6) p value thresholds. Known variants are in blue, novel ones in red. For each lead variant, the nearest gene is presented, with the color corresponding to its position to the lead variant: exonic (red), 3′-UTR (green), intronic (blue), intergenic including up- and downstream, exonic and intronic non-coding RNA (gray). Nearest genes for more than one locus are denoted with an asterisk (∗).
(B) Circos heatmap showing the betas of lead variants in African, Asian, and European ancestry meta-analyses, as well as the transancestral meta-analysis. Differences between the height-unadjusted (model 1) and -adjusted (model 2) meta-analysis are also shown.
(C) Bar plot of the genetic correlation coefficient (ρgenetic) of the height-unadjusted and -adjusted head size GWAS with the height GWAS, with their accompanying 95% confidence intervals.