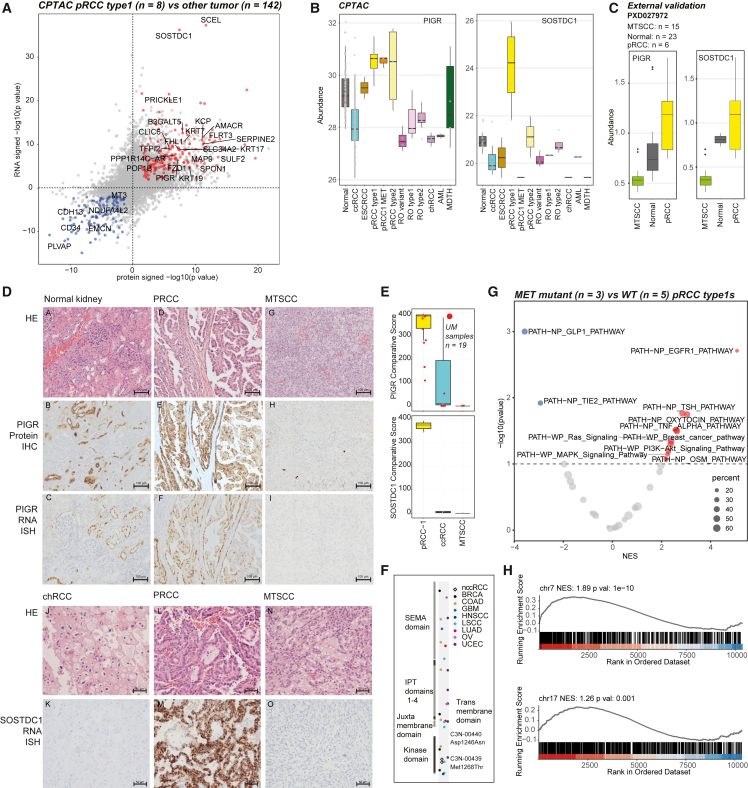
Figure 6.
Proteogenomic biomarkers that distinguish pRCC from MTSCC
(A) Significantly differential events (abs(log2fc) > 2 and q < 0.05) in protein expression (x axis) and RNA expression (y axis) between pRCC type 1 and other tumors.
(B) Specificity of pRCC type 1 protein markers PIGR and SOSTDC1.
(C) Expression of pRCC type 1 protein markers PIGR and SOSTDC1 in the proteomics data from Xu et al.23 (PXD027972).
(D) H&E, protein IHC, and RNA-ISH images (top to bottom) of biomarker PIGR in normal kidney tissue, pRCC, MTSCC tumors (upper panels from left to right) and SOSTDC1 in chRCC, pRCC, and MTSCC (lower panels from left to right).
(E) RNA-ISH comparative scores of PIGR and SOSTDC1 in different tumor types. Red points represent external University of Michigan samples.
(F) Location of missense mutations in MET across TCGA cohorts are colored on the MET protein domain diagram.
(G) PTM-SEA analysis shows pathways such as EGFR are significantly enriched with increased phosphorylation in MET mutant pRCC samples.
(H) Enrichment in chromosomes 7 and 17 gene sets are tested with protein expression difference between chromosome 7 gain and no gain non-ccRCC sample groups.