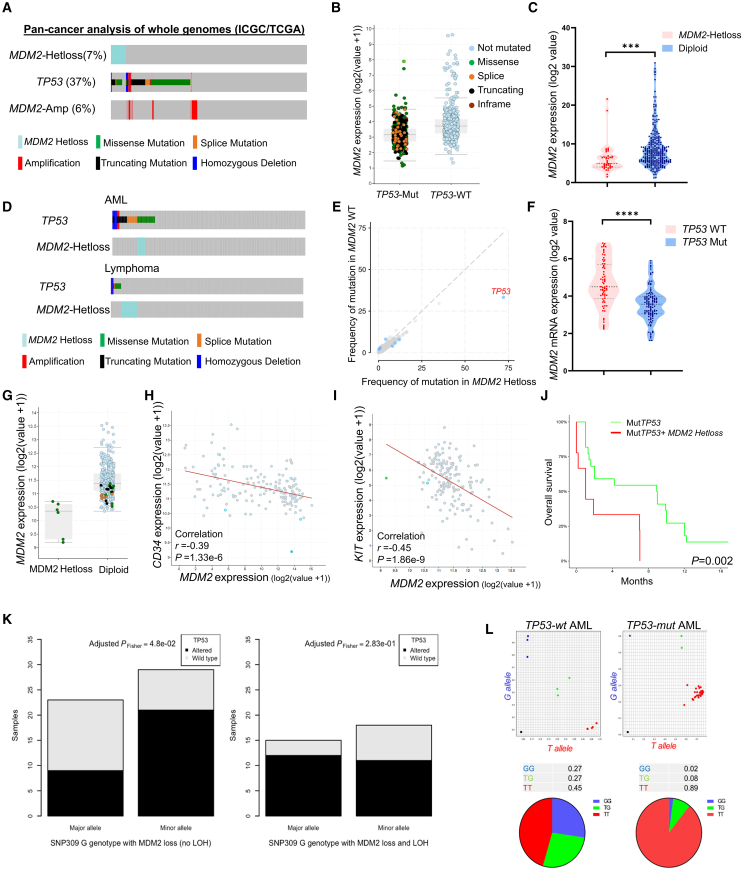
Figure 1.
Analysis of TP53 mutations, MDM2 alterations, and MDM2 expression patterns in cancer
(A) Frequency of TP53 mutations and MDM2 copy number alterations across various cancer types using whole-genome analysis of ICGC/TCGA Pan-Cancer data.
(B) Expression of MDM2 in TP53–wild-type (WT) and TP53-mutant (Mut) cases in pan-cancer analysis of whole genomes.
(C) Relative expression of MDM2 mRNA based on MDM2 allele status pan-cancer analysis of whole genomes, ∗∗∗p < 0.001.
(D) Frequency of TP53 mutations and MDM2 copy number alterations in patients with AML and lymphoma, sourced from cBioPortal for Cancer Genomics.
(E) Frequency of mutations in different genes categorized by MDM2 WT and MDM2 LOH, retrieved from cBioPortal.
(F) Relative expression of MDM2 based on TP53 status using data from the Cancer Cell Line Encyclopedia, ∗∗∗∗p < 0.0001.
(G) MDM2 expression in AML samples, highlighting cases with mutant TP53: green for missense, brown for splice, and black for truncation mutations.
(H and I) Scatter diagram graphs the correlation of CD34 and KIT versus MDM2 at the mRNA levels in patients with AML (TCGA).
(J) Overall survival of p53 mutant AML patients with or without MDM2 LOH.
(K) Bar plot displaying the distribution of the G allele on the major or minor allele in germline T/G cases and in the context of somatic MDM2 loss, depending on TP53 mutational status (altered or wild type) and the presence or absence of the T allele (no LOH; left) or its absence (LOH; right). This demonstrates a significant enrichment for the G allele on the minor allele in altered TP53 cases without LOH (adjusted PFisher = 4.8e−2).
(L) Allele-specific expression analysis of GG (blue), TG (green), and TT (red) in indicated patient samples.