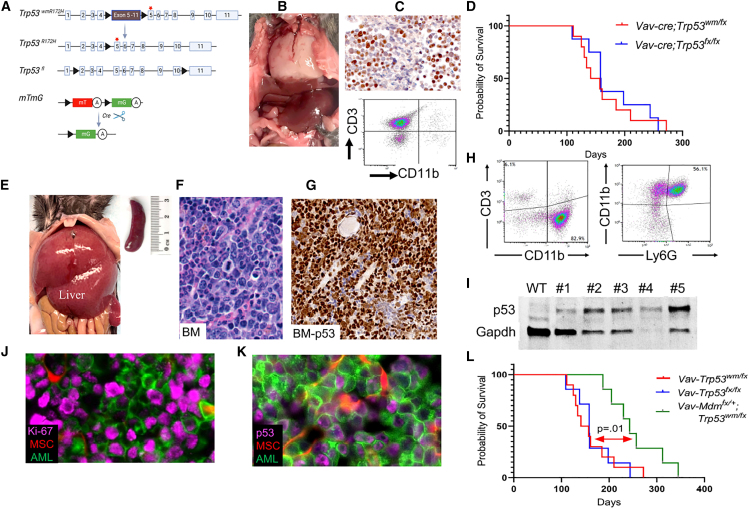
Figure 2.
Overview of alleles and pathological findings in a novel p53 mutant AML mouse model
(A) Schematic representation of the genetic modifications in the mouse model. The activation of Cre recombinase under the Vav1 promoter leads to deletion of wild-type Trp53 cDNA flanked by loxP sites (black triangles) resulting in a Trp53-mut allele. The diagram also shows the recombination of the mTmG reporter allele, facilitating GFP expression in hematopoietic cells, with “mT” for membranous tomato, “mG” for membranous GFP, and “A” indicating a polyadenylation sequence. The red star marks the Trp53 R172H mutation on exon 5.
(B) Image of a white thoracic lymphoma mass in a Vav-Cre;Trp53fl/wmR172H mouse.
(C) Immunohistochemistry for p53, showing nuclear accumulation in lymphoma cells. Flow cytometry analysis identifying a distinct CD3+ T lymphocyte population, with absence of CD11b+ myeloid cells.
(D) Kaplan-Meier survival curve with a p value of 0.2 from the log rank (Mantel-Cox) test, showing the overall survival rate of the mice.
(E) Representative image of AML infiltration in liver and spleen of moribund Vav-Cre;Mdm2+/fl;Trp53fl/wmR172H mice.
(F) Hematoxylin and eosin (H&E)-stained bone marrow (BM) section from the AML mouse depicted in (J), showing characteristic features of AML.
(G) Immunostaining for p53 protein, visualized as brown staining, demonstrates the presence of p53 in leukemia cells within bone marrow (BM).
(H) Flow cytometry analysis of CD3, CD11b, and Ly6G markers in the bone marrow of AML mice.
(I) Western blot analysis of p53 protein expression in BM samples comparing p53 protein levels in p53 wild-type control and five AML samples.
(J) Immunofluorescence analysis of Ki-67 in BM. MSCs, mesenchymal stromal cells.
(K) Mutant p53 accumulation in GFP-positive AML cells in BM sections.
(L) Kaplan-Meier analysis of overall survival of the indicated mice. p = 0.01, log rank (Mantel-Cox) test.