Figure 3.
Clonal evolution and generation of a p53 mutant clonal hematopoiesis mouse model
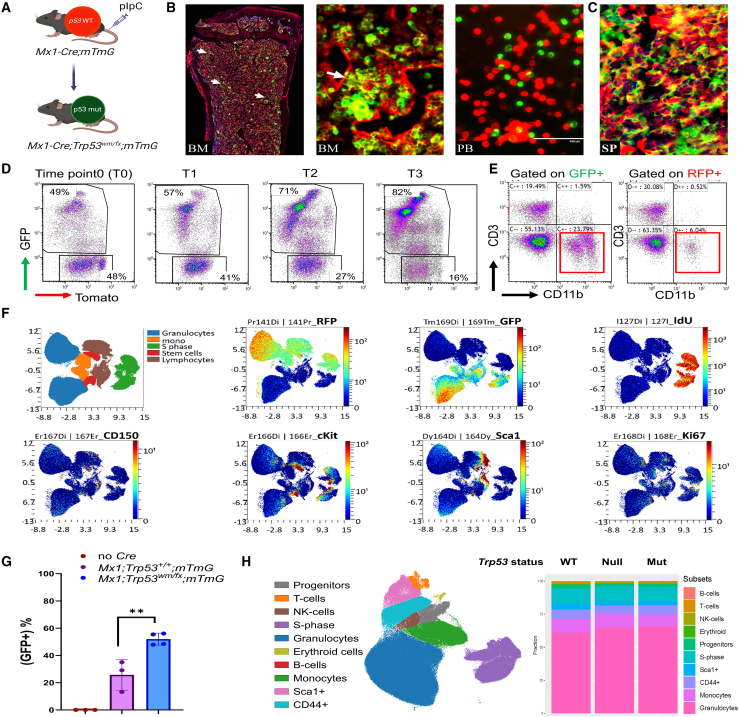
(A) The schematic outlines the creation of the Trp53R172H clonal hematopoiesis model in adult HSCs. Following pIpC injection, Cre recombinase expressed under the Mx1-Cre promoter induces recombination in hematopoietic cells, leading to a fluorescence switch in the mTmG allele from red (tdTomato) to green (GFP), simultaneously converting wild-type p53 to its mutant form.
(B and C) (B) Direct fluorescence image of the bone marrow of Mx1-Cre;Trp53wmR172 H/fl;mTmG mouse showing the distribution of GFP+ cells in the bone marrow (BM), peripheral blood (PB), and (C) spleen (SP). High magnification displaying a colony of GFP+ cells marked by white arrow.
(D) Flow cytometry plots showing the expression of GFP and Tomato in the peripheral blood of Mx1-Cre;Trp53wmR172 H/fl;mTmG mice at different time points.
(E) Flow cytometry plots of CD3 and CD11b expression gated on GFP or RFP in (D).
(F) tSNE plots representing a CyTOF analysis of the indicated marker distribution in BM.
(G) Graph showing the percentage of GFP+ cells in the peripheral blood of indicated mice over 3 months after pIpC injection. The data are presented as mean ± standard deviation.
(H) UMAP plot showing the cell subsets detected in pooled BM samples isolated from Mx1-Cre;mTmG (p53 wild-type), Mx1-Cre;Trp53fl/fl;mTmG (p53 null), and Mx1-Cre;Trp53wmR172 H/fl;mTmG (p53 mutant) mice subjected to UMAP dimension reduction. Each linage is marked by colors indicating cell type. The stacked bar plots summarize the subset frequencies in indicated genotypes.