Figure 3.
Heart disease promotes inflammatory microvascular dysfunction in the right atrium
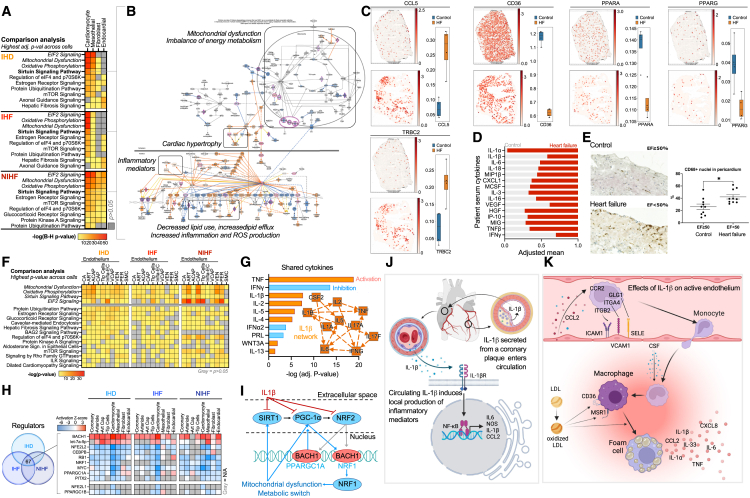
(A) Top 10 canonical pathways with the highest significance score (B–H adjusted p value <0.05 for all) by IPA’s comparison analysis across the main cell types, as described in STAR Methods, in IHD (N = 11), IHF (N = 11), and NIHF (N = 3) against control (N = 6). Full table and details available at http://compbio2.mit.edu/scheart/.
(B) Sirtuin signaling pathway from IPA for CMs in NIHF against control as a representative for all three groups. Blue indicates inhibition of the molecule, orange/red activation. Purple highlight marks significantly differentially expressed molecules in the dataset. Full version of the pathway figure can be found in Figure S2A.
(C) Spatial expression (Visium) of TRBC, CCL5, CD36, PPARA, and PPARG in a control and heart failure (HF) sample. For each gene, quantitation of the signal is shown across control (n = 4) and HF (n = 4) sections (right) and in one representative section (left), as described in STAR Methods. Whiskers show the maximum and minimum values, except for outliers (more than 1.5 times the interquartile).
(D) Scaled (0–1) mean of serum cytokines in patients with HF (N = 6) and controls (N = 7). No statistical significance.
(E) Quantitation of CD68+ tissue MPs in pericardium of patients with HF (N = 10) and control group (N = 10) (∗p < 0.0104). A representative immunohistostaining image provided for both groups, showing MPs in brown.
(F) Top 10 canonical pathways with the highest significance score (Fisher’s exact test, p < 0.05 for all) across vascular cells within each group: IHD, IHF, and NIHF. Top 10 pathways for each condition were selected and p value is presented across all conditions.
(G) Top shared cytokines in IPA’s Upstream Regulator analysis between IHD, IHF, NIHF, and disturbed flow in HUVECs dataset (N = 3 for d-flow and for control). IL-1β network from 2-h time point by IPA’s machine-learning-based graphical summary is a representative for all four time points (2, 8, 14, and 32 h of IL-1β treatment compared to control [N = 3] in HAECs).
(H) Top overlapping upstream regulators (Fisher’s exact test p < 0.05 for all) with similar activity patterns from IPA (microRNAs and transcriptional regulators only) for IHD, IHF, and NIHF in main cell types and four EC subtypes. Red is for predicted activity and blue for inhibition.
(I) Depiction of the connections between the potential mediators of the mitochondrial and metabolic changes.
(J) Overview of IL-1β signaling.
(K) Effects of IL-1β on activated endothelium.
(J) and (K) were created with BioRender.com.